[English] 日本語
 Yorodumi
Yorodumi- PDB-3cti: RELAXATION MATRIX REFINEMENT OF THE SOLUTION STRUCTURE OF SQUASH ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3cti | ||||||
|---|---|---|---|---|---|---|---|
| Title | RELAXATION MATRIX REFINEMENT OF THE SOLUTION STRUCTURE OF SQUASH TRYPSIN INHIBITOR | ||||||
 Components Components | TRYPSIN INHIBITOR | ||||||
 Keywords Keywords | PROTEINASE INHIBITOR (TRYPSIN) | ||||||
| Function / homology | Proteinase inhibitor I7, squash / Squash family serine protease inhibitor / Squash family of serine protease inhibitors signature. / Plant trypsin inhibitors / Proteinase/amylase inhibitor domain superfamily / serine-type endopeptidase inhibitor activity / extracellular region / Trypsin inhibitor 1 Function and homology information Function and homology information | ||||||
| Biological species |  Cucurbita maxima (winter squash) Cucurbita maxima (winter squash) | ||||||
| Method | SOLUTION NMR | ||||||
 Authors Authors | Nilges, M. / Habazettl, J. / Bruenger, A.T. / Holak, T.A. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1991 Journal: J.Mol.Biol. / Year: 1991Title: Relaxation matrix refinement of the solution structure of squash trypsin inhibitor. Authors: Nilges, M. / Habazettl, J. / Brunger, A.T. / Holak, T.A. #1:  Journal: J.Mol.Biol. / Year: 1989 Journal: J.Mol.Biol. / Year: 1989Title: Determination of the Complete Three-Dimensional Structure of the Trypsin Inhibitor from Squash Seeds in Aqueous Solution by Nuclear Magnetic Resonance and a Combination of Distance Geometry ...Title: Determination of the Complete Three-Dimensional Structure of the Trypsin Inhibitor from Squash Seeds in Aqueous Solution by Nuclear Magnetic Resonance and a Combination of Distance Geometry and Dynamical Simulated Annealing Authors: Holak, T.A. / Gondol, D. / Otlewski, J. / Wilusz, T. #2:  Journal: J.Mol.Biol. / Year: 1989 Journal: J.Mol.Biol. / Year: 1989Title: Nuclear Magnetic Resonance Solution and X-Ray Structures of Squash Trypsin Inhibitor Exhibit the Same Conformation of the Proteinase Binding Loop Authors: Holak, T.A. / Bode, W. / Huber, R. / Otlewski, J. / Wilusz, T. #3:  Journal: FEBS Lett. / Year: 1990 Journal: FEBS Lett. / Year: 1990Title: 1H NMR Assignments of Sidechain Conformations in Proteins Using a High-Dimensional Potential in the Simulated Annealing Calculations Authors: Habazettl, J. / Cieslar, C. / Oschkinat, H. / Holak, T.A. | ||||||
| History |
|
- Structure visualization
Structure visualization
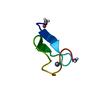
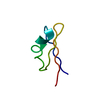
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3cti.cif.gz 3cti.cif.gz | 58.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3cti.ent.gz pdb3cti.ent.gz | 45.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3cti.json.gz 3cti.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ct/3cti https://data.pdbj.org/pub/pdb/validation_reports/ct/3cti ftp://data.pdbj.org/pub/pdb/validation_reports/ct/3cti ftp://data.pdbj.org/pub/pdb/validation_reports/ct/3cti | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 3279.919 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Cucurbita maxima (winter squash) / References: UniProt: P01074 Cucurbita maxima (winter squash) / References: UniProt: P01074 |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Crystal grow | *PLUS Method: other / Details: NMR |
|---|
- Processing
Processing
| Software |
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software |
| ||||||||||
| NMR ensemble | Conformers submitted total number: 6 |
 Movie
Movie Controller
Controller






 PDBj
PDBj X-PLOR
X-PLOR