[English] 日本語
 Yorodumi
Yorodumi- PDB-2zjj: Crystal structure of the human BACE1 catalytic domain in complex ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2zjj | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the human BACE1 catalytic domain in complex with 4-(4-fluoro-benzyl)-piperazine-2-carboxylic acid (2-mercapto-ethyl)-amide | ||||||
 Components Components | Beta-secretase 1 | ||||||
 Keywords Keywords | HYDROLASE / BACE1 / small-molecule inhibitor / Tethering / Aspartyl protease / Glycoprotein / Membrane / Protease / Transmembrane / Zymogen | ||||||
| Function / homology |  Function and homology information Function and homology informationmemapsin 2 / Golgi-associated vesicle lumen / signaling receptor ligand precursor processing / beta-aspartyl-peptidase activity / amyloid precursor protein catabolic process / amyloid-beta formation / membrane protein ectodomain proteolysis / detection of mechanical stimulus involved in sensory perception of pain / amyloid-beta metabolic process / cellular response to manganese ion ...memapsin 2 / Golgi-associated vesicle lumen / signaling receptor ligand precursor processing / beta-aspartyl-peptidase activity / amyloid precursor protein catabolic process / amyloid-beta formation / membrane protein ectodomain proteolysis / detection of mechanical stimulus involved in sensory perception of pain / amyloid-beta metabolic process / cellular response to manganese ion / prepulse inhibition / protein serine/threonine kinase binding / cellular response to copper ion / presynaptic modulation of chemical synaptic transmission / multivesicular body / hippocampal mossy fiber to CA3 synapse / response to lead ion / trans-Golgi network / recycling endosome / protein processing / positive regulation of neuron apoptotic process / cellular response to amyloid-beta / late endosome / synaptic vesicle / peptidase activity / amyloid-beta binding / endopeptidase activity / amyloid fibril formation / lysosome / aspartic-type endopeptidase activity / early endosome / endosome membrane / endosome / membrane raft / Amyloid fiber formation / endoplasmic reticulum lumen / axon / neuronal cell body / dendrite / Golgi apparatus / enzyme binding / cell surface / proteolysis / membrane / plasma membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å | ||||||
 Authors Authors | Randal, M. / Lam, M.B. / Romanowski, M.J. | ||||||
 Citation Citation |  Journal: To be Published Journal: To be PublishedTitle: Fragment-based discovery of novel BACE1 inhibitors using Tethering technology Authors: Yang, W. / Fucini, R.V. / Fahr, B.T. / Randal, M. / Lind, K.E. / Lam, M.B. / Lu, W. / Lu, Y. / Cary, D.R. / Romanowski, M.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2zjj.cif.gz 2zjj.cif.gz | 91.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2zjj.ent.gz pdb2zjj.ent.gz | 67.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2zjj.json.gz 2zjj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2zjj_validation.pdf.gz 2zjj_validation.pdf.gz | 758.3 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2zjj_full_validation.pdf.gz 2zjj_full_validation.pdf.gz | 760.3 KB | Display | |
| Data in XML |  2zjj_validation.xml.gz 2zjj_validation.xml.gz | 16.5 KB | Display | |
| Data in CIF |  2zjj_validation.cif.gz 2zjj_validation.cif.gz | 23.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zj/2zjj https://data.pdbj.org/pub/pdb/validation_reports/zj/2zjj ftp://data.pdbj.org/pub/pdb/validation_reports/zj/2zjj ftp://data.pdbj.org/pub/pdb/validation_reports/zj/2zjj | HTTPS FTP |
-Related structure data
| Related structure data |  2zjhC  2zjiC  2zjkC  2zjlC  2zjmC  2zjnC  2p8hS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 45124.789 Da / Num. of mol.: 1 / Fragment: BACE1 catalytic domain, UNP residues 43-446 / Mutation: K75A, E77A, T231C Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: BACE1 / Plasmid: PET11A / Production host: Homo sapiens (human) / Gene: BACE1 / Plasmid: PET11A / Production host:  |
|---|---|
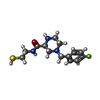
| #2: Chemical | ChemComp-F1J / ( |
| #3: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.45 Å3/Da / Density % sol: 49.75 % |
|---|---|
| Crystal grow | Temperature: 295 K / Method: vapor diffusion, sitting drop / pH: 8 Details: 0.1M imidazole pH 8.0, 0.2M Ca(OAC)2, 10% PEG 8000, VAPOR DIFFUSION, SITTING DROP, temperature 295K |
-Data collection
| Diffraction | Mean temperature: 180 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 5.0.1 / Wavelength: 0.98 Å / Beamline: 5.0.1 / Wavelength: 0.98 Å |
| Detector | Type: ADSC QUANTUM 210 / Detector: CCD / Date: Jun 4, 2004 |
| Radiation | Monochromator: Si(220) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.98 Å / Relative weight: 1 |
| Reflection | Resolution: 2.2→20 Å / Num. obs: 18114 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2P8H Resolution: 2.2→20 Å / Cor.coef. Fo:Fc: 0.927 / Cor.coef. Fo:Fc free: 0.906 / SU B: 7.696 / SU ML: 0.194 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / ESU R: 0.548 / ESU R Free: 0.298 / Stereochemistry target values: MAXIMUM LIKELIHOOD
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 34.255 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.2→2.276 Å / Total num. of bins used: 15
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: 13.672 Å / Origin y: 37.891 Å / Origin z: 25.471 Å
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj