[English] 日本語
 Yorodumi
Yorodumi- PDB-2w14: High-resolution crystal structure of the P-I snake venom metallop... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2w14 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | High-resolution crystal structure of the P-I snake venom metalloproteinase BaP1 in complex with a peptidomimetic: insights into inhibitor binding | |||||||||
 Components Components | ZINC METALLOPROTEINASE BAP1 | |||||||||
 Keywords Keywords | HYDROLASE/INHIBITOR / HYDROLASE INHIBITOR COMPLEX / METAL-BINDING / ZINC-DEPENDING / METALLOPROTEASE / METALLOPROTEINASE-INHIBITOR COMPLEX / ZINC / TOXIN / SECRETED / PROTEASE / HYDROLASE / P-I SNAKE VENOM METALLOPROTEINASE / METZINCIN / CHEMOTAXIS / ADAMALYSIN / ENDOPEPTIDASE / ALPHA-BETA PROTEIN / MATRIXMETALLOPROTEINASE / TUMOR NECROSIS FACTOR ALPHA CONVERTING ENZYME / PYRROLIDONE CARBOXYLIC ACID / HYDROLASE-INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases / metalloendopeptidase activity / chemotaxis / toxin activity / proteolysis / extracellular region / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  BOTHROPS ASPER (terciopelo) BOTHROPS ASPER (terciopelo) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.08 Å MOLECULAR REPLACEMENT / Resolution: 1.08 Å | |||||||||
 Authors Authors | Lingott, T.J. / Schleberger, C. / Gutierrez, J.M. / Merfort, I. | |||||||||
 Citation Citation |  Journal: Biochemistry / Year: 2009 Journal: Biochemistry / Year: 2009Title: High-Resolution Crystal Structure of the Snake Venom Metalloproteinase Bap1 Complexed with a Peptidomimetic: Insight Into Inhibitor Binding. Authors: Lingott, T.J. / Schleberger, C. / Gutierrez, J.M. / Merfort, I. #1:  Journal: Protein Sci. / Year: 2003 Journal: Protein Sci. / Year: 2003Title: Amino Acid Sequence and Crystal Structure of Bap1, a Metalloproteinase from Bothrops Asper Snake Venom that Exerts Multiple Tissue-Damaging Activities. Authors: Watanabe, L. / Shannon, J.D. / Valente, R.H. / Rucavado, A. / Alape-Giron, A. / Kamiguti, A.S. / Theakston, R.D.G. / Fox, J.W. / Gutierrez, J.M. / Arni, R.K. | |||||||||
| History |
| |||||||||
| Remark 650 | HELIX DETERMINATION METHOD: AUTHOR PROVIDED. | |||||||||
| Remark 700 | SHEET DETERMINATION METHOD: AUTHOR PROVIDED. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2w14.cif.gz 2w14.cif.gz | 116 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2w14.ent.gz pdb2w14.ent.gz | 88.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2w14.json.gz 2w14.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/w1/2w14 https://data.pdbj.org/pub/pdb/validation_reports/w1/2w14 ftp://data.pdbj.org/pub/pdb/validation_reports/w1/2w14 ftp://data.pdbj.org/pub/pdb/validation_reports/w1/2w14 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2w12C  2w13SC  2w15C C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 22770.740 Da / Num. of mol.: 1 / Fragment: RESIDUES 193-394 / Source method: isolated from a natural source / Source: (natural)  BOTHROPS ASPER (terciopelo) / Secretion: VENOM BOTHROPS ASPER (terciopelo) / Secretion: VENOMReferences: UniProt: P83512, Hydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases |
|---|
-Non-polymers , 5 types, 394 molecules 
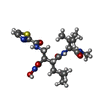







| #2: Chemical | ChemComp-ZN / |
|---|---|
| #3: Chemical | ChemComp-WR2 / ( |
| #4: Chemical | ChemComp-GOL / |
| #5: Chemical | ChemComp-IMD / |
| #6: Water | ChemComp-HOH / |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.07 Å3/Da / Density % sol: 40.7 % / Description: NONE |
|---|---|
| Crystal grow | pH: 8 / Details: PEG 8000, IMIDAZOLE, pH 8.0 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  BESSY BESSY  / Beamline: 14.1 / Wavelength: 0.91841 / Beamline: 14.1 / Wavelength: 0.91841 |
| Detector | Type: MARRESEARCH / Detector: CCD |
| Radiation | Monochromator: KMC-1, DOUBLE CRYSTAL / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.91841 Å / Relative weight: 1 |
| Reflection | Resolution: 1.08→20 Å / Num. obs: 77288 / % possible obs: 94.6 % / Observed criterion σ(I): 4 / Redundancy: 7.4 % / Biso Wilson estimate: 8.67 Å2 / Rmerge(I) obs: 0.07 / Net I/σ(I): 17.6 |
| Reflection shell | Resolution: 1.08→1.11 Å / Rmerge(I) obs: 0.33 / Mean I/σ(I) obs: 4.2 / % possible all: 64.6 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2W13 Resolution: 1.08→18.22 Å / Cor.coef. Fo:Fc: 0.975 / Cor.coef. Fo:Fc free: 0.974 / SU B: 0.674 / SU ML: 0.016 / Cross valid method: THROUGHOUT / ESU R: 0.025 / ESU R Free: 0.026 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. ATOMS OF LOOP 159-162 ARE MODELED (SEMI- OCCUPIED) IN TWO DIFFERENT CONFORMATIONS.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 9.79 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.08→18.22 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj










