[English] 日本語
 Yorodumi
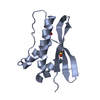
Yorodumi- PDB-2vpd: Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signa... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2vpd | ||||||
|---|---|---|---|---|---|---|---|
| Title | Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | GENE REGULATION / WNT SIGNALING PATHWAY / WNT SIGNALING COMPLEX / CHROMOSOMAL REARRANGEMENT / SIGNALING PROTEIN / BCL9 HD1 DOMAIN / HPYGO1 PHD DOMAIN / PROTO-ONCOGENE / PHOSPHOPROTEIN / HISTONE H3K4ME2 / ZINC / NUCLEUS / ZINC-FINGER / METAL-BINDING | ||||||
| Function / homology |  Function and homology information Function and homology informationmyotube differentiation involved in skeletal muscle regeneration / spermatid nucleus differentiation / beta-catenin-TCF complex / cis-Golgi network / histone H3K4me3 reader activity / myoblast differentiation / skeletal muscle cell differentiation / somatic stem cell population maintenance / sarcoplasm / canonical Wnt signaling pathway ...myotube differentiation involved in skeletal muscle regeneration / spermatid nucleus differentiation / beta-catenin-TCF complex / cis-Golgi network / histone H3K4me3 reader activity / myoblast differentiation / skeletal muscle cell differentiation / somatic stem cell population maintenance / sarcoplasm / canonical Wnt signaling pathway / protein localization to nucleus / hematopoietic progenitor cell differentiation / kidney development / Deactivation of the beta-catenin transactivating complex / Formation of the beta-catenin:TCF transactivating complex / beta-catenin binding / transcription by RNA polymerase II / transcription coactivator activity / positive regulation of transcription by RNA polymerase II / zinc ion binding / nucleoplasm Similarity search - Function | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.77 Å MOLECULAR REPLACEMENT / Resolution: 2.77 Å | ||||||
 Authors Authors | Fiedler, M. / Sanchez-Barrena, M.J. / Nekrasov, M. / Mieszczanek, J. / Rybin, V. / Muller, J. / Evans, P. / Bienz, M. | ||||||
 Citation Citation |  Journal: Mol.Cell / Year: 2008 Journal: Mol.Cell / Year: 2008Title: Decoding of Methylated Histone H3 Tail by the Pygo- Bcl9 Wnt Signaling Complex. Authors: Fiedler, M. / Sanchez-Barrena, M.J. / Nekrasov, M. / Mieszczanek, J. / Rybin, V. / Muller, J. / Evans, P. / Bienz, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2vpd.cif.gz 2vpd.cif.gz | 50.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2vpd.ent.gz pdb2vpd.ent.gz | 35.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2vpd.json.gz 2vpd.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vp/2vpd https://data.pdbj.org/pub/pdb/validation_reports/vp/2vpd ftp://data.pdbj.org/pub/pdb/validation_reports/vp/2vpd ftp://data.pdbj.org/pub/pdb/validation_reports/vp/2vpd | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2vp7C  2vpbSC  2vpeC  2vpgC C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 7229.039 Da / Num. of mol.: 2 / Fragment: PHD DOMAIN, RESIDUES 333-398 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Production host: HOMO SAPIENS (human) / Production host:  #2: Protein/peptide | Mass: 3758.405 Da / Num. of mol.: 2 / Fragment: HD1 DOMAIN, RESIDUES 174-205 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Production host: HOMO SAPIENS (human) / Production host:  #3: Chemical | ChemComp-ZN / #4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.1 Å3/Da / Density % sol: 60.34 % / Description: NONE |
|---|---|
| Crystal grow | Temperature: 292 K / Method: vapor diffusion, hanging drop Details: HANGING-DROP VAPOUR-DIFFUSION METHOD AT 19 CELSIUS. CRYSTALLIZATION CONDITIONS: 1.3M (NH4)2SO4, 100MM TRIS PH6.5, 200MM NACL |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-3 / Wavelength: 0.931 / Beamline: ID14-3 / Wavelength: 0.931 |
| Detector | Type: ADSC CCD / Detector: CCD / Date: Jun 16, 2007 / Details: TOROIDAL MIRROR |
| Radiation | Monochromator: DIAMOND (111), GE(220) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.931 Å / Relative weight: 1 |
| Reflection | Resolution: 2.77→31.45 Å / Num. obs: 30994 / % possible obs: 99.7 % / Observed criterion σ(I): 2 / Redundancy: 7 % / Rmerge(I) obs: 0.06 / Net I/σ(I): 22.5 |
| Reflection shell | Resolution: 2.77→2.92 Å / Redundancy: 6.9 % / Rmerge(I) obs: 0.38 / Mean I/σ(I) obs: 4.2 / % possible all: 99.7 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2VPB Resolution: 2.77→66.67 Å / Cor.coef. Fo:Fc: 0.933 / Cor.coef. Fo:Fc free: 0.905 / SU B: 16.298 / SU ML: 0.316 / Cross valid method: THROUGHOUT / ESU R: 0.682 / ESU R Free: 0.339 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. THE PHD RESIDUE S362 PRESENTS AN UNUSUAL CONFORMATION SINCE IT IS NEXT TO C363, WHICH IS PART OF THE ZN 2 BINDING SITE
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 64.85 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.77→66.67 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj

