+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2pfo | ||||||
|---|---|---|---|---|---|---|---|
| Title | DNA Polymerase lambda in complex with DNA and dUPNPP | ||||||
 Components Components |
| ||||||
 Keywords Keywords | transferase / lyase/DNA / DNA polymerase / DNA Repair / phosphoryl transfer reaction / manganese / lyase-DNA COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationDNA biosynthetic process / Lyases; Carbon-oxygen lyases; Other carbon-oxygen lyases / 5'-deoxyribose-5-phosphate lyase activity / somatic hypermutation of immunoglobulin genes / base-excision repair, gap-filling / nucleotide-excision repair / Nonhomologous End-Joining (NHEJ) / double-strand break repair via homologous recombination / double-strand break repair via nonhomologous end joining / site of double-strand break ...DNA biosynthetic process / Lyases; Carbon-oxygen lyases; Other carbon-oxygen lyases / 5'-deoxyribose-5-phosphate lyase activity / somatic hypermutation of immunoglobulin genes / base-excision repair, gap-filling / nucleotide-excision repair / Nonhomologous End-Joining (NHEJ) / double-strand break repair via homologous recombination / double-strand break repair via nonhomologous end joining / site of double-strand break / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / DNA replication / DNA binding / nucleoplasm / metal ion binding / nucleus Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | ||||||
 Authors Authors | Garcia-Diaz, M. / Bebenek, K. / Krahn, J.M. / Pedersen, L.C. / Kunkel, T.A. | ||||||
 Citation Citation |  Journal: DNA Repair / Year: 2007 Journal: DNA Repair / Year: 2007Title: Role of the catalytic metal during polymerization by DNA polymerase lambda. Authors: Garcia-Diaz, M. / Bebenek, K. / Krahn, J.M. / Pedersen, L.C. / Kunkel, T.A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2pfo.cif.gz 2pfo.cif.gz | 103.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2pfo.ent.gz pdb2pfo.ent.gz | 71.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2pfo.json.gz 2pfo.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2pfo_validation.pdf.gz 2pfo_validation.pdf.gz | 804 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2pfo_full_validation.pdf.gz 2pfo_full_validation.pdf.gz | 810.7 KB | Display | |
| Data in XML |  2pfo_validation.xml.gz 2pfo_validation.xml.gz | 19.2 KB | Display | |
| Data in CIF |  2pfo_validation.cif.gz 2pfo_validation.cif.gz | 28.5 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pf/2pfo https://data.pdbj.org/pub/pdb/validation_reports/pf/2pfo ftp://data.pdbj.org/pub/pdb/validation_reports/pf/2pfo ftp://data.pdbj.org/pub/pdb/validation_reports/pf/2pfo | HTTPS FTP |
-Related structure data
| Related structure data |  2pfnC  2pfpC  2pfqC  1xsnS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-DNA chain , 3 types, 3 molecules TPD
| #1: DNA chain | Mass: 3374.210 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source |
|---|---|
| #2: DNA chain | Mass: 1793.219 Da / Num. of mol.: 1 / Source method: obtained synthetically |
| #3: DNA chain | Mass: 1191.818 Da / Num. of mol.: 1 / Source method: obtained synthetically |
-Protein , 1 types, 1 molecules A
| #4: Protein | Mass: 37447.688 Da / Num. of mol.: 1 / Mutation: C543A Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: POLL / Production host: Homo sapiens (human) / Gene: POLL / Production host:  References: UniProt: Q9UGP5, DNA-directed DNA polymerase, Lyases; Carbon-oxygen lyases; Other carbon-oxygen lyases |
|---|
-Non-polymers , 6 types, 337 molecules 


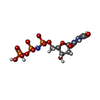







| #5: Chemical | ChemComp-MG / | ||||||
|---|---|---|---|---|---|---|---|
| #6: Chemical | ChemComp-MN / | ||||||
| #7: Chemical | ChemComp-NA / #8: Chemical | ChemComp-DUP / | #9: Chemical | ChemComp-EDO / | #10: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.74 Å3/Da / Density % sol: 55.1 % | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 278 K / Method: vapor diffusion, hanging drop / pH: 5.5 Details: 10-20% 2-propanol, 0.2 M sodium citrate and 0.1M sodium cacodylate , pH 5.5, VAPOR DIFFUSION, HANGING DROP, temperature 278K | ||||||||||||||||||||||||||||
| Components of the solutions |
|
-Data collection
| Diffraction | Mean temperature: 95 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU / Wavelength: 1.5418 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: RIGAKU SATURN 92 / Detector: CCD / Date: May 16, 2005 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2→50 Å / Num. obs: 33210 / % possible obs: 98.9 % / Redundancy: 5.4 % / Rmerge(I) obs: 0.058 / Χ2: 1.025 / Net I/σ(I): 23.1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1XSN Resolution: 2→50 Å / σ(F): 0
| ||||||||||||||||||||||||||||||||
| Solvent computation | Bsol: 52.333 Å2 | ||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 38.346 Å2
| ||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→50 Å
| ||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj











































