[English] 日本語
 Yorodumi
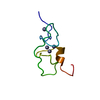
Yorodumi- PDB-2myh: Omega-Tbo-IT1: selective inhibitor of insect calcium channels iso... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2myh | ||||||
|---|---|---|---|---|---|---|---|
| Title | Omega-Tbo-IT1: selective inhibitor of insect calcium channels isolated from Tibellus oblongus spider venom | ||||||
 Components Components | Omega-Tbo-IT1 toxin | ||||||
 Keywords Keywords | TOXIN / cystine knot inhibitor / ICK | ||||||
| Function / homology | host cell presynaptic membrane / calcium channel regulator activity / toxin activity / extracellular region / Omega-phylotoxin-To1a Function and homology information Function and homology information | ||||||
| Biological species |  Tibellus oblongus (spider) Tibellus oblongus (spider) | ||||||
| Method | SOLUTION NMR / simulated annealing, torsion angle dynamics | ||||||
 Authors Authors | Altukhov, D. / Bozin, T. / Bocharov, E. / Kozlov, S. / Mikov, A. | ||||||
 Citation Citation |  Journal: Sci Rep / Year: 2015 Journal: Sci Rep / Year: 2015Title: omega-Tbo-IT1-New Inhibitor of Insect Calcium Channels Isolated from Spider Venom. Authors: Mikov, A.N. / Fedorova, I.M. / Potapieva, N.N. / Maleeva, E.E. / Andreev, Y.A. / Zaitsev, A.V. / Kim, K.K. / Bocharov, E.V. / Bozin, T.N. / Altukhov, D.A. / Lipkin, A.V. / Kozlov, S.A. / ...Authors: Mikov, A.N. / Fedorova, I.M. / Potapieva, N.N. / Maleeva, E.E. / Andreev, Y.A. / Zaitsev, A.V. / Kim, K.K. / Bocharov, E.V. / Bozin, T.N. / Altukhov, D.A. / Lipkin, A.V. / Kozlov, S.A. / Tikhonov, D.B. / Grishin, E.V. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2myh.cif.gz 2myh.cif.gz | 267.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2myh.ent.gz pdb2myh.ent.gz | 223.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2myh.json.gz 2myh.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/my/2myh https://data.pdbj.org/pub/pdb/validation_reports/my/2myh ftp://data.pdbj.org/pub/pdb/validation_reports/my/2myh ftp://data.pdbj.org/pub/pdb/validation_reports/my/2myh | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 4348.183 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Tibellus oblongus (spider) / Plasmid: pET-32-b(+) / Production host: Tibellus oblongus (spider) / Plasmid: pET-32-b(+) / Production host:  |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR Details: TWO-DIMENSIONAL 1H-1H DQF-COSY, TOCSY (80-MS MIXING TIME), NOESY (120-MS MIXING TIME) AND 1H-13C HSQC (AT NATURAL ABUNDANCE) NMR SPECTRA OF 1.5 MM RECOMBINANT OMEGA-TBO-IT1 TOXIN ...Details: TWO-DIMENSIONAL 1H-1H DQF-COSY, TOCSY (80-MS MIXING TIME), NOESY (120-MS MIXING TIME) AND 1H-13C HSQC (AT NATURAL ABUNDANCE) NMR SPECTRA OF 1.5 MM RECOMBINANT OMEGA-TBO-IT1 TOXIN (SOLUBILIZED IN H2O OR D2O WITH 20 MM PHOSPHATE BUFFER, PH 5.8) WERE ACQUIRED AT 12C AND 20C ON NMR SPECTROMETER VARIAN NMR SYSTEM 700 MHZ. | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| |||||||||
| Sample conditions | Ionic strength: 0.06 / pH: 5.8 / Pressure: ambient / Temperature: 285 K |
-NMR measurement
| NMR spectrometer | Type: Varian Uniform NMR System 700 / Manufacturer: Varian / Model: Uniform NMR System 700 / Field strength: 700 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing, torsion angle dynamics / Software ordinal: 1 | ||||||||||||||||||||||||||||||||||||
| NMR constraints | NOE constraints total: 422 / NOE intraresidue total count: 249 / NOE long range total count: 49 / NOE medium range total count: 21 / NOE sequential total count: 103 / Disulfide bond constraints total count: 24 / Hydrogen bond constraints total count: 88 / Protein chi angle constraints total count: 89 / Protein other angle constraints total count: 0 / Protein phi angle constraints total count: 89 / Protein psi angle constraints total count: 2 | ||||||||||||||||||||||||||||||||||||
| NMR representative | Selection criteria: fewest violations | ||||||||||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: target function / Conformers calculated total number: 100 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller









 PDBj
PDBj HSQC
HSQC