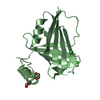
登録情報 データベース : PDB / ID : 2mvxタイトル Atomic-resolution 3D structure of amyloid-beta fibrils: the Osaka mutation Amyloid beta A4 protein キーワード / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Homo sapiens (ヒト)手法 / データ登録者 Schuetz, A.K. / Vagt, T. / Huber, M. / Ovchinnikova, O.Y. / Cadalbert, R. / Wall, J. / Guentert, P. / Bockmann, A. / Glockshuber, R. / Meier, B.H. ジャーナル : Angew.Chem.Int.Ed.Engl. / 年 : 2015タイトル : Atomic-Resolution Three-Dimensional Structure of Amyloid beta Fibrils Bearing the Osaka Mutation.著者 : Schutz, A.K. / Vagt, T. / Huber, M. / Ovchinnikova, O.Y. / Cadalbert, R. / Wall, J. / Guntert, P. / Bockmann, A. / Glockshuber, R. / Meier, B.H. 履歴 登録 2014年10月17日 登録サイト / 処理サイト 改定 1.0 2014年11月26日 Provider / タイプ 改定 1.1 2015年1月21日 Group 改定 1.2 2024年5月1日 Group / Database referencesカテゴリ chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_nmr_spectrometer / struct_ref_seq_dif Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_nmr_spectrometer.model / _struct_ref_seq_dif.details
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト) データ登録者
データ登録者 引用
引用 ジャーナル: Angew.Chem.Int.Ed.Engl. / 年: 2015
ジャーナル: Angew.Chem.Int.Ed.Engl. / 年: 2015 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 2mvx.cif.gz
2mvx.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb2mvx.ent.gz
pdb2mvx.ent.gz PDB形式
PDB形式 2mvx.json.gz
2mvx.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 2mvx_validation.pdf.gz
2mvx_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 2mvx_full_validation.pdf.gz
2mvx_full_validation.pdf.gz 2mvx_validation.xml.gz
2mvx_validation.xml.gz 2mvx_validation.cif.gz
2mvx_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/mv/2mvx
https://data.pdbj.org/pub/pdb/validation_reports/mv/2mvx ftp://data.pdbj.org/pub/pdb/validation_reports/mv/2mvx
ftp://data.pdbj.org/pub/pdb/validation_reports/mv/2mvx リンク
リンク 集合体
集合体
 要素
要素 Homo sapiens (ヒト) / 遺伝子: A4, AD1, APP / プラスミド: pET T7 / 発現宿主:
Homo sapiens (ヒト) / 遺伝子: A4, AD1, APP / プラスミド: pET T7 / 発現宿主: 
 試料調製
試料調製 解析
解析 ムービー
ムービー コントローラー
コントローラー












 PDBj
PDBj














 CYANA
CYANA