[English] 日本語
 Yorodumi
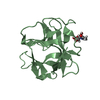
Yorodumi- PDB-2mm3: Solution NMR structure of the ternary complex of human ileal bile... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2mm3 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution NMR structure of the ternary complex of human ileal bile acid-binding protein with glycocholate and glycochenodeoxycholate | ||||||
 Components Components | Gastrotropin | ||||||
 Keywords Keywords | LIPID BINDING PROTEIN / lipid-binding protein / orthogonal beta sheets / positive binding cooperativity / site-selectivity / enterohepatic circulation | ||||||
| Function / homology |  Function and homology information Function and homology informationNR1H2 & NR1H3 regulate gene expression to control bile acid homeostasis / Triglyceride catabolism / fatty acid transport / Recycling of bile acids and salts / fatty acid binding / lipid metabolic process / negative regulation of cell population proliferation / lipid binding / nucleus / membrane ...NR1H2 & NR1H3 regulate gene expression to control bile acid homeostasis / Triglyceride catabolism / fatty acid transport / Recycling of bile acids and salts / fatty acid binding / lipid metabolic process / negative regulation of cell population proliferation / lipid binding / nucleus / membrane / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | SOLUTION NMR / simulated annealing, torsion angle dynamics | ||||||
| Model details | closest to the average, model9 | ||||||
 Authors Authors | Horvath, G. / Egyed, O. / Bencsura, A. / Simon, A. / Tochtrop, G.P. / DeKoster, G.T. / Covey, D.F. / Cistola, D.P. / Toke, O. | ||||||
 Citation Citation |  Journal: FEBS J. / Year: 2016 Journal: FEBS J. / Year: 2016Title: Structural determinants of ligand binding in the ternary complex of human ileal bile acid binding protein with glycocholate and glycochenodeoxycholate obtained from solution NMR Authors: Horvath, G. / Bencsura, A. / Simon, A. / Tochtrop, G.P. / DeKoster, G.T. / Covey, D.F. / Cistola, D.P. / Toke, O. #1: Journal: Biochemistry / Year: 2006 Title: Determinants of cooperativity and site selectivity in human ileal bile acid binding protein. Authors: Toke, O. / Monsey, J.D. / DeKoster, G.T. / Tochtrop, G.P. / Tang, C. / Cistola, D.P. #2: Journal: J.Am.Chem.Soc. / Year: 2004 Title: A single hydroxyl group governs ligand site selectivity in human ileal bile acid binding protein. Authors: Tochtrop, G.P. / DeKoster, G.T. / Covey, D.F. / Cistola, D.P. #3: Journal: Biochemistry / Year: 2007 Title: Kinetic mechanism of ligand binding in human ileal bile acid binding protein as determined by stopped-flow fluorescence analysis. Authors: Toke, O. / Monsey, J.D. / Cistola, D.P. #4: Journal: Biochemistry / Year: 2012 Title: Internal motions and exchange processes in human ileal bile acid binding protein as studied by backbone (15)N nuclear magnetic resonance spectroscopy. Authors: Horvath, G. / Kiraly, P. / Tarkanyi, G. / Toke, O. #5:  Journal: Biochemistry / Year: 2012 Journal: Biochemistry / Year: 2012Title: Internal motions and exchange processes in human ileal bile acid binding protein as studied by backbone (15)N nuclear magnetic resonance spectroscopy Authors: Horvath, G. / Kiraly, P. / Tarkanyi, G. / Toke, O. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2mm3.cif.gz 2mm3.cif.gz | 457.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2mm3.ent.gz pdb2mm3.ent.gz | 390.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2mm3.json.gz 2mm3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2mm3_validation.pdf.gz 2mm3_validation.pdf.gz | 666.3 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2mm3_full_validation.pdf.gz 2mm3_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  2mm3_validation.xml.gz 2mm3_validation.xml.gz | 131.4 KB | Display | |
| Data in CIF |  2mm3_validation.cif.gz 2mm3_validation.cif.gz | 94.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mm/2mm3 https://data.pdbj.org/pub/pdb/validation_reports/mm/2mm3 ftp://data.pdbj.org/pub/pdb/validation_reports/mm/2mm3 ftp://data.pdbj.org/pub/pdb/validation_reports/mm/2mm3 | HTTPS FTP |
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 14258.038 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: FABP6 / Production host: Homo sapiens (human) / Gene: FABP6 / Production host:  |
|---|---|
| #2: Chemical | ChemComp-GCH / |
| #3: Chemical | ChemComp-CHO / |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sample conditions | Ionic strength: 70 / pH: 6.3 / Pressure: ambient / Temperature: 293 K |
-NMR measurement
| NMR spectrometer | Type: Varian Varian NMR System / Manufacturer: Varian / Model: Varian NMR System / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing, torsion angle dynamics / Software ordinal: 1 | ||||||||||||||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 100 / Conformers submitted total number: 10 / Representative conformer: 9 |
 Movie
Movie Controller
Controller











 PDBj
PDBj






 HSQC
HSQC