| 1 | 1.3-2.2 mM 5'-R(*GP*AP*GP*CP*AP*GP*CP*AP*UP*CP*GP*UP*CP*GP*GP*CP*UP*GP*CP*UP*CP*A)-3', 1.3-2.2 mM [U-15N] 5'-R(*GP*CP*GP*GP*CP*AP*GP*UP*UP*GP*AP*CP*UP*AP*CP*UP*GP*UP*CP*GP*C)-3', 10 mM [U-2H] Tris, 50 mM sodium chloride, 0.05 mM sodium azide, 5 mM magnesium chloride, 90% H2O/10% D2O | 90% H2O/10% D2O| 2 | 1.5 mM 5'-R(*GP*AP*GP*CP*AP*GP*CP*AP*UP*CP*GP*UP*CP*GP*GP*CP*UP*GP*CP*UP*CP*A)-3', 1.5 mM [U-13C; U-15N] 5'-R(*GP*CP*GP*GP*CP*AP*GP*UP*UP*GP*AP*CP*UP*AP*CP*UP*GP*UP*CP*GP*C)-3', 10 mM [U-2H] Tris, 50 mM sodium chloride, 0.05 mM sodium azide, 5 mM magnesium chloride, 100% D2O | 100% D2O| 3 | 1.4 mM [U-15N] 5'-R(*GP*AP*GP*CP*AP*GP*CP*AP*UP*CP*GP*UP*CP*GP*GP*CP*UP*GP*CP*UP*CP*A)-3', 1.4 mM 5'-R(*GP*CP*GP*GP*CP*AP*GP*UP*UP*GP*AP*CP*UP*AP*CP*UP*GP*UP*CP*GP*C)-3', 10 mM [U-2H] Tris, 50 mM sodium chloride, 0.05 mM sodium azide, 5 mM magnesium chloride, 90% H2O/10% D2O | 90% H2O/10% D2O| 4 | 1.7 mM [U-13C; U-15N] 5'-R(*GP*AP*GP*CP*AP*GP*CP*AP*UP*CP*GP*UP*CP*GP*GP*CP*UP*GP*CP*UP*CP*A)-3', 1.7 mM 5'-R(*GP*CP*GP*GP*CP*AP*GP*UP*UP*GP*AP*CP*UP*AP*CP*UP*GP*UP*CP*GP*C)-3', 10 mM [U-2H] Tris, 50 mM sodium chloride, 0.05 mM sodium azide, 5 mM magnesium chloride, 100% D2O | 100% D2O| 5 | 1.4 mM [U-13C; U-15N]-Gua 5'-R(*GP*AP*GP*CP*AP*GP*CP*AP*UP*CP*GP*UP*CP*GP*GP*CP*UP*GP*CP*UP*CP*A)-3', 1.4 mM 5'-R(*GP*CP*GP*GP*CP*AP*GP*UP*UP*GP*AP*CP*UP*AP*CP*UP*GP*UP*CP*GP*C)-3', 10 mM [U-2H] Tris, 50 mM sodium chloride, 0.05 mM sodium azide, 5 mM magnesium chloride, 100% D2O | 100% D2O| 6 | 1.4 mM [U-13C; U-15N]-Gua 5'-R(*GP*AP*GP*CP*AP*GP*CP*AP*UP*CP*GP*UP*CP*GP*GP*CP*UP*GP*CP*UP*CP*A)-3', 1.4 mM 5'-R(*GP*CP*GP*GP*CP*AP*GP*UP*UP*GP*AP*CP*UP*AP*CP*UP*GP*UP*CP*GP*C)-3', 10 mM [U-2H] Tris, 50 mM sodium chloride, 0.05 mM sodium azide, 5 mM magnesium chloride, 100% D2O | 100% D2O| 7 | 1.4 mM [U-15N] 5'-R(*GP*AP*GP*CP*AP*GP*CP*AP*UP*CP*GP*UP*CP*GP*GP*CP*UP*GP*CP*UP*CP*A)-3', 1.4 mM 5'-R(*GP*CP*GP*GP*CP*AP*GP*UP*UP*GP*AP*CP*UP*AP*CP*UP*GP*UP*CP*GP*C)-3', 10 mM [U-2H] Tris, 50 mM sodium chloride, 0.05 mM sodium azide, 5 mM magnesium chloride, 100% D2O | 100% D2O| 8 | 1.3 mM 5'-R(*GP*AP*GP*CP*AP*GP*CP*AP*UP*CP*GP*UP*CP*GP*GP*CP*UP*GP*CP*UP*CP*A)-3', 1.3 mM [U-15N] 5'-R(*GP*CP*GP*GP*CP*AP*GP*UP*UP*GP*AP*CP*UP*AP*CP*UP*GP*UP*CP*GP*C)-3', 10 mM [U-2H] Tris, 50 mM sodium chloride, 0.05 mM sodium azide, 5 mM magnesium chloride, 100% D2O | 100% D2O| 9 | 1.7 mM [U-13C; U-15N] 5'-R(*GP*AP*GP*CP*AP*GP*CP*AP*UP*CP*GP*UP*CP*GP*GP*CP*UP*GP*CP*UP*CP*A)-3', 1.7 mM 5'-R(*GP*CP*GP*GP*CP*AP*GP*UP*UP*GP*AP*CP*UP*AP*CP*UP*GP*UP*CP*GP*C)-3', 10 mM [U-2H] Tris, 50 mM sodium chloride, 0.05 mM sodium azide, 5 mM magnesium chloride, 90% H2O/10% D2O | 90% H2O/10% D2O| 10 | 1.5 mM 5'-R(*GP*AP*GP*CP*AP*GP*CP*AP*UP*CP*GP*UP*CP*GP*GP*CP*UP*GP*CP*UP*CP*A)-3', 1.5 mM [U-13C; U-15N] 5'-R(*GP*CP*GP*GP*CP*AP*GP*UP*UP*GP*AP*CP*UP*AP*CP*UP*GP*UP*CP*GP*C)-3', 10 mM [U-2H] Tris, 50 mM sodium chloride, 0.05 mM sodium azide, 5 mM magnesium chloride, 90% H2O/10% D2O | 90% H2O/10% D2O| 11 | 0.2-0.3 mM [U-15N] 5'-R(*GP*AP*GP*CP*AP*GP*CP*AP*UP*CP*GP*UP*CP*GP*GP*CP*UP*GP*CP*UP*CP*A)-3', 0.2-0.3 mM [U-15N] 5'-R(*GP*CP*GP*GP*CP*AP*GP*UP*UP*GP*AP*CP*UP*AP*CP*UP*GP*UP*CP*GP*C)-3', 10 mM [U-2H] Tris, 50 mM sodium chloride, 0.05 mM sodium azide, 5 mM magnesium chloride, 14.6-20.5 mg/mL Pf1 phage, 90% H2O/10% D2O | 90% H2O/10% D2O | | | | | | | | | | |
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Neurospora (fungus)
Neurospora (fungus) Authors
Authors Citation
Citation Journal: Biochemistry / Year: 2014
Journal: Biochemistry / Year: 2014 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2mi0.cif.gz
2mi0.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2mi0.ent.gz
pdb2mi0.ent.gz PDB format
PDB format 2mi0.json.gz
2mi0.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 2mi0_validation.pdf.gz
2mi0_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 2mi0_full_validation.pdf.gz
2mi0_full_validation.pdf.gz 2mi0_validation.xml.gz
2mi0_validation.xml.gz 2mi0_validation.cif.gz
2mi0_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/mi/2mi0
https://data.pdbj.org/pub/pdb/validation_reports/mi/2mi0 ftp://data.pdbj.org/pub/pdb/validation_reports/mi/2mi0
ftp://data.pdbj.org/pub/pdb/validation_reports/mi/2mi0 Links
Links Assembly
Assembly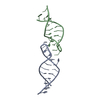
 Components
Components Neurospora (fungus)
Neurospora (fungus) Neurospora (fungus)
Neurospora (fungus) Sample preparation
Sample preparation Movie
Movie Controller
Controller







 PDBj
PDBj





























 HSQC
HSQC