[English] 日本語
 Yorodumi
Yorodumi- PDB-2mco: Structural studies on dinuclear ruthenium(II) complexes that bind... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2mco | ||||||
|---|---|---|---|---|---|---|---|
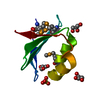
| Title | Structural studies on dinuclear ruthenium(II) complexes that bind diastereoselectively to an anti-parallel folded human telomere sequence | ||||||
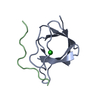
 Components Components | human telomere quadruplex | ||||||
 Keywords Keywords | DNA / quadruplex / complex / ruthenium | ||||||
| Function / homology | Chem-2FJ / DNA / DNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / torsion angle dynamics | ||||||
| Model details | lowest energy, model1 | ||||||
 Authors Authors | Williamson, M.P. / Wilson, T. / Thomas, J.A. / Felix, V. / Costa, P.J. | ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2013 Journal: J.Med.Chem. / Year: 2013Title: Structural Studies on Dinuclear Ruthenium(II) Complexes That Bind Diastereoselectively to an Antiparallel Folded Human Telomere Sequence. Authors: Wilson, T. / Costa, P.J. / Felix, V. / Williamson, M.P. / Thomas, J.A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2mco.cif.gz 2mco.cif.gz | 102.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2mco.ent.gz pdb2mco.ent.gz | 72.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2mco.json.gz 2mco.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mc/2mco https://data.pdbj.org/pub/pdb/validation_reports/mc/2mco ftp://data.pdbj.org/pub/pdb/validation_reports/mc/2mco ftp://data.pdbj.org/pub/pdb/validation_reports/mc/2mco | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2mccC C: citing same article ( |
|---|---|
| Similar structure data | |
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 6983.497 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: standard solid phase synthesis, hplc purified |
|---|---|
| #2: Chemical | ChemComp-2FJ / |
| #3: Chemical |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR Details: Complex of LL-[{Ru(bipy)2}2tppz]4+ bound to human telomere sequence quadruplex DNA (basket). Structures used NMR intermolecular restraints followed by unrestrained MD. The 5 structures in ...Details: Complex of LL-[{Ru(bipy)2}2tppz]4+ bound to human telomere sequence quadruplex DNA (basket). Structures used NMR intermolecular restraints followed by unrestrained MD. The 5 structures in the ensemble are representatives of the 5 most populated clusters from a 50 ns simulation. Simulation had explicit solvent plus counterions, which are removed in these files except for two sodiums in the middle of the quadruplex. | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 0.3 mM human telomere quadruplex, 0.3 mM LL-(Ru[bipy]2)tppz4+, 50 mM sodium chloride, 100% D2O Solvent system: 100% D2O | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||
| Sample conditions | Ionic strength: 50 / pH: 7 / Pressure: ambient / Temperature: 298 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: torsion angle dynamics / Software ordinal: 1 | ||||||||||||
| NMR constraints | NOE constraints total: 13 / NOE intraresidue total count: 0 / NOE long range total count: 0 / NOE medium range total count: 0 / NOE sequential total count: 0 | ||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||
| NMR ensemble | Conformer selection criteria: all calculated structures submitted Conformers calculated total number: 5 / Conformers submitted total number: 5 / Maximum lower distance constraint violation: 0.5 Å / Maximum upper distance constraint violation: 0.5 Å | ||||||||||||
| NMR ensemble rms | Distance rms dev: 0.1 Å / Distance rms dev error: 0.05 Å |
 Movie
Movie Controller
Controller






 PDBj
PDBj









































 HSQC
HSQC