[English] 日本語
 Yorodumi
Yorodumi- PDB-2lqz: Structure of the RNA claw of the DNA packaging motor of bacteriop... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2lqz | ||||||
|---|---|---|---|---|---|---|---|
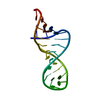
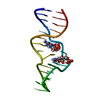
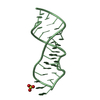
| Title | Structure of the RNA claw of the DNA packaging motor of bacteriophage 29 | ||||||
 Components Components | RNA (27-MER) | ||||||
 Keywords Keywords | RNA / bulge | ||||||
| Function / homology | RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
| Model details | closest to the average, model 1 | ||||||
 Authors Authors | Harjes, E.J. / Matsuo, H.J. / Kitamura, A.J. | ||||||
 Citation Citation |  Journal: Nucleic Acids Res. / Year: 2012 Journal: Nucleic Acids Res. / Year: 2012Title: Structure of the RNA claw of the DNA packaging motor of bacteriophage 29. Authors: Harjes, E. / Kitamura, A. / Zhao, W. / Morais, M.C. / Jardine, P.J. / Grimes, S. / Matsuo, H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2lqz.cif.gz 2lqz.cif.gz | 151.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2lqz.ent.gz pdb2lqz.ent.gz | 127.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2lqz.json.gz 2lqz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lq/2lqz https://data.pdbj.org/pub/pdb/validation_reports/lq/2lqz ftp://data.pdbj.org/pub/pdb/validation_reports/lq/2lqz ftp://data.pdbj.org/pub/pdb/validation_reports/lq/2lqz | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 8586.128 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 300 uM [U-100% 13C; U-100% 15N] RNA (27-MER), 90% H2O/10% D2O Solvent system: 90% H2O/10% D2O |
|---|---|
| Sample | Conc.: 300 uM / Component: RNA (27-MER)-1 / Isotopic labeling: [U-100% 13C; U-100% 15N] |
| Sample conditions | Ionic strength: 50 / pH: 6.5 / Pressure: ambient / Temperature: 298 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software | Name: CNS / Developer: Brunger A. T. et.al. / Classification: refinement |
|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 |
| NMR constraints | NA alpha-angle constraints total count: 12 / NA beta-angle constraints total count: 13 / NA chi-angle constraints total count: 12 / NA delta-angle constraints total count: 16 / NA epsilon-angle constraints total count: 12 / NA gamma-angle constraints total count: 13 / NA other-angle constraints total count: 29 / NA sugar pucker constraints total count: 99 / NOE constraints total: 429 / NOE intraresidue total count: 136 / NOE long range total count: 68 / NOE medium range total count: 33 / NOE sequential total count: 192 |
| NMR representative | Selection criteria: closest to the average |
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 50 / Conformers submitted total number: 9 |
 Movie
Movie Controller
Controller








 PDBj
PDBj





























 HSQC
HSQC