| Entry | Database: PDB / ID: 2kcn
|
|---|
| Title | Solution structure of the antifungal protein PAF from Penicillium chrysogenum |
|---|
 Components Components | Antifungal protein |
|---|
 Keywords Keywords | ANTIFUNGAL PROTEIN / antifungal protein PAF |
|---|
| Function / homology |  Function and homology information Function and homology information
Antifungal protein domain / Antifungal protein / Antifungal protein domain superfamily / Antifungal protein / OB fold (Dihydrolipoamide Acetyltransferase, E2P) / Beta Barrel / Mainly BetaSimilarity search - Domain/homology |
|---|
| Biological species |  Penicillium chrysogenum (fungus) Penicillium chrysogenum (fungus) |
|---|
| Method | SOLUTION NMR / torsion angle dynamics |
|---|
| Model details | closest to the average, model 4 |
|---|
 Authors Authors | Batta, G. / Barna, T. / Gaspari, Z. / Sandor, S. / Kover, K.E. / Binder, U. / Sarg, B. / Kaiserer, L. / Chhillar, A.K. / Eigentler, A. ...Batta, G. / Barna, T. / Gaspari, Z. / Sandor, S. / Kover, K.E. / Binder, U. / Sarg, B. / Kaiserer, L. / Chhillar, A.K. / Eigentler, A. / Leiter, E. / Hegedus, N. / Pocsi, I. / Lindner, H. / Marx, F. |
|---|
 Citation Citation |  Journal: Febs J. / Year: 2009 Journal: Febs J. / Year: 2009
Title: Functional aspects of the solution structure and dynamics of PAF--a highly-stable antifungal protein from Penicillium chrysogenum
Authors: Batta, G. / Barna, T. / Gaspari, Z. / Sandor, S. / Kover, K.E. / Binder, U. / Sarg, B. / Kaiserer, L. / Chhillar, A.K. / Eigentler, A. / Leiter, E. / Hegedus, N. / Pocsi, I. / Lindner, H. / Marx, F. |
|---|
| History | | Deposition | Dec 23, 2008 | Deposition site: BMRB / Processing site: PDBJ |
|---|
| Revision 1.0 | Jul 21, 2009 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jul 13, 2011 | Group: Version format compliance |
|---|
| Revision 1.2 | Feb 26, 2020 | Group: Derived calculations / Other
Category: pdbx_database_status / pdbx_struct_assembly / pdbx_struct_oper_list
Item: _pdbx_database_status.status_code_cs |
|---|
| Revision 1.3 | Jun 14, 2023 | Group: Database references / Other / Category: database_2 / pdbx_database_status
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_database_status.status_code_nmr_data |
|---|
| Revision 1.4 | May 15, 2024 | Group: Data collection / Database references / Category: chem_comp_atom / chem_comp_bond / database_2 / Item: _database_2.pdbx_DOI |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Penicillium chrysogenum (fungus)
Penicillium chrysogenum (fungus) Authors
Authors Citation
Citation Journal: Febs J. / Year: 2009
Journal: Febs J. / Year: 2009 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2kcn.cif.gz
2kcn.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2kcn.ent.gz
pdb2kcn.ent.gz PDB format
PDB format 2kcn.json.gz
2kcn.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/kc/2kcn
https://data.pdbj.org/pub/pdb/validation_reports/kc/2kcn ftp://data.pdbj.org/pub/pdb/validation_reports/kc/2kcn
ftp://data.pdbj.org/pub/pdb/validation_reports/kc/2kcn Links
Links Assembly
Assembly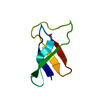
 Components
Components Penicillium chrysogenum (fungus) / Gene: paf / References: UniProt: Q01701
Penicillium chrysogenum (fungus) / Gene: paf / References: UniProt: Q01701 Sample preparation
Sample preparation Processing
Processing Movie
Movie Controller
Controller











 PDBj
PDBj HSQC
HSQC