+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2k76 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution structure of a paralog-specific Mena binder by NMR | ||||||
 Components Components | pGolemi | ||||||
 Keywords Keywords | DE NOVO PROTEIN / protein design / miniature protein / aPP / beta-hairpin / ActA homolog | ||||||
| Method | SOLUTION NMR / simulated annealing, torsion angle dynamics | ||||||
 Authors Authors | Link, N.M. / Hunke, C. / Eichler, J. / Bayer, P. | ||||||
 Citation Citation |  Journal: Biol.Chem. / Year: 2009 Journal: Biol.Chem. / Year: 2009Title: The solution structure of pGolemi, a high affinity Mena EVH1 binding miniature protein, suggests explanations for paralog-specific binding to Ena/VASP homology (EVH) 1 domains. Authors: Link, N.M. / Hunke, C. / Mueller, J.W. / Eichler, J. / Bayer, P. #1:  Journal: J.Am.Chem.Soc. / Year: 2004 Journal: J.Am.Chem.Soc. / Year: 2004Title: High Affinity, Paralog-Specific Recognition of the Mena EVH1 domain by a miniature protein Authors: Golemi-Kotra, D. / Mahaffy, R. / Footer, M.J. / Holtzman, J.H. / Pollard, T.D. / Theriot, J.A. / Schepartz, A. #2:  Journal: Biochemistry / Year: 2007 Journal: Biochemistry / Year: 2007Title: Miniature Protein Ligands for EVH1 Domains: Interplay between Affinity, Specificity, and Cell Motility Authors: Holtzman, J.H. / Woronowicz, K. / Golemi-Kotra, D. / Schepartz, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2k76.cif.gz 2k76.cif.gz | 98.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2k76.ent.gz pdb2k76.ent.gz | 80.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2k76.json.gz 2k76.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/k7/2k76 https://data.pdbj.org/pub/pdb/validation_reports/k7/2k76 ftp://data.pdbj.org/pub/pdb/validation_reports/k7/2k76 ftp://data.pdbj.org/pub/pdb/validation_reports/k7/2k76 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 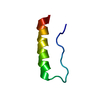
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 3459.831 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: Solid-phase peptide synthesis. The original sequence comes from Meleagris gallopavo (Wild Turkey) |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 1 mM pGolemi, 90% H2O, 10% D2O / Solvent system: 90% H2O/10% D2O |
|---|---|
| Sample | Conc.: 1 mM / Component: pGolemi |
| Sample conditions | Ionic strength: 45 / pH: 7 / Pressure: ambient / Temperature: 300 K |
-NMR measurement
| NMR spectrometer | Type: Bruker Avance / Manufacturer: Bruker / Model: DRX / Field strength: 700 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing, torsion angle dynamics / Software ordinal: 1 | ||||||||||||||||||||
| NMR constraints | NOE constraints total: 373 / NOE intraresidue total count: 203 / NOE long range total count: 12 / NOE medium range total count: 63 / NOE sequential total count: 95 / Hydrogen bond constraints total count: 26 / Protein other angle constraints total count: 50 | ||||||||||||||||||||
| NMR representative | Selection criteria: fewest violations | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: lowest energy / Conformers calculated total number: 200 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller










 PDBj
PDBj
 X-PLOR NIH
X-PLOR NIH