+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2igu | ||||||
|---|---|---|---|---|---|---|---|
| Title | Deamidated analogue of ImI Conotoxin | ||||||
 Components Components | Alpha-conotoxin ImI | ||||||
 Keywords Keywords | TOXIN / Conotoxin / disulfide linkage / ribbon conformation | ||||||
| Function / homology | Conotoxin, alpha-type, conserved site / Alpha-conotoxin family signature. / host cell postsynaptic membrane / acetylcholine receptor inhibitor activity / ion channel regulator activity / toxin activity / extracellular region / Alpha-conotoxin ImI Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / Energy Minization, Molecular Dynamics, Simulated Annealing | ||||||
| Model type details | minimized average | ||||||
 Authors Authors | Kini, R.M. / Kang, T.S. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2007 Journal: Biochemistry / Year: 2007Title: Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins Authors: Kang, T.S. / Talley, T.T. / Jois, S.D. / Taylor, P. / Kini, R.M. #1: Journal: Angew.Chem.Int.Ed.Engl. / Year: 2005 Title: Effect of C-terminal amidation on folding and disulfide-pairing of alpha-conotoxin ImI Authors: Kang, T.S. / Vivekanandan, S. / Jois, S.D. / Kini, R.M. | ||||||
| History |
|
- Structure visualization
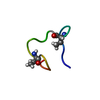
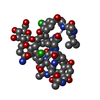
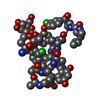
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2igu.cif.gz 2igu.cif.gz | 46.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2igu.ent.gz pdb2igu.ent.gz | 32.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2igu.json.gz 2igu.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ig/2igu https://data.pdbj.org/pub/pdb/validation_reports/ig/2igu ftp://data.pdbj.org/pub/pdb/validation_reports/ig/2igu ftp://data.pdbj.org/pub/pdb/validation_reports/ig/2igu | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2ifiC  2ifjC  2ifzC  2ih6C  2ih7C  2ihaC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 1358.594 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: Chemically synthesized using Fmoc solid phase peptide synthesis. References: UniProt: P50983*PLUS |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 2mM peptide sample, pH 3.0; 10% D2O, 90% H2O / Solvent system: 10% D2O, 90% H2O |
|---|---|
| Sample conditions | Ionic strength: 2mM / pH: 3 / Pressure: ambient / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Bruker DRX / Manufacturer: Bruker / Model: DRX / Field strength: 500 MHz |
|---|
- Processing
Processing
| NMR software |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: Energy Minization, Molecular Dynamics, Simulated Annealing Software ordinal: 1 | |||||||||||||||
| NMR representative | Selection criteria: minimized average structure | |||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 200 / Conformers submitted total number: 14 |
 Movie
Movie Controller
Controller





 PDBj
PDBj