+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 2hqh | ||||||
|---|---|---|---|---|---|---|---|
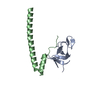
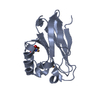
| タイトル | Crystal structure of p150Glued and CLIP-170 | ||||||
 要素 要素 |
| ||||||
 キーワード キーワード | STRUCTURAL PROTEIN / PROTEIN BINDING / beta/beta structure / zinc finger motif | ||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報cell cortex region / centriolar subdistal appendage / positive regulation of neuromuscular junction development / centriole-centriole cohesion / microtubule anchoring at centrosome / maintenance of synapse structure / ventral spinal cord development / microtubule plus-end / nuclear membrane disassembly / positive regulation of microtubule nucleation ...cell cortex region / centriolar subdistal appendage / positive regulation of neuromuscular junction development / centriole-centriole cohesion / microtubule anchoring at centrosome / maintenance of synapse structure / ventral spinal cord development / microtubule plus-end / nuclear membrane disassembly / positive regulation of microtubule nucleation / XBP1(S) activates chaperone genes / melanosome transport / microtubule bundle formation / microtubule plus-end binding / non-motile cilium assembly / dynein complex / retrograde transport, endosome to Golgi / COPI-independent Golgi-to-ER retrograde traffic / Signaling by LTK in cancer / intermediate filament / microtubule associated complex / neuromuscular process / nuclear migration / neuromuscular junction development / cell leading edge / motor behavior / establishment of mitotic spindle orientation / RHO GTPases activate IQGAPs / intercellular bridge / COPI-mediated anterograde transport / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / ruffle / cytoplasmic microtubule organization / positive regulation of microtubule polymerization / neuron projection maintenance / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / centriole / Loss of Nlp from mitotic centrosomes / Loss of proteins required for interphase microtubule organization from the centrosome / Recruitment of mitotic centrosome proteins and complexes / MHC class II antigen presentation / tubulin binding / Recruitment of NuMA to mitotic centrosomes / Anchoring of the basal body to the plasma membrane / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / Resolution of Sister Chromatid Cohesion / regulation of mitotic spindle organization / cytoplasmic vesicle membrane / AURKA Activation by TPX2 / RHO GTPases Activate Formins / neuron cellular homeostasis / kinetochore / tau protein binding / spindle / spindle pole / mitotic spindle / Separation of Sister Chromatids / Signaling by ALK fusions and activated point mutants / Regulation of PLK1 Activity at G2/M Transition / nuclear envelope / nervous system development / mitotic cell cycle / microtubule cytoskeleton / cell cortex / microtubule binding / microtubule / neuron projection / cilium / ciliary basal body / axon / cell division / neuronal cell body / centrosome / protein kinase binding / zinc ion binding / identical protein binding / nucleus / membrane / plasma membrane / cytoplasm / cytosol 類似検索 - 分子機能 | ||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | ||||||
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  多波長異常分散 / 解像度: 1.8 Å 多波長異常分散 / 解像度: 1.8 Å | ||||||
 データ登録者 データ登録者 | Hayashi, I. / Ikura, M. | ||||||
 引用 引用 |  ジャーナル: Nat.Struct.Mol.Biol. / 年: 2007 ジャーナル: Nat.Struct.Mol.Biol. / 年: 2007タイトル: CLIP170 autoinhibition mimics intermolecular interactions with p150Glued or EB1. 著者: Hayashi, I. / Plevin, M.J. / Ikura, M. | ||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  2hqh.cif.gz 2hqh.cif.gz | 95.6 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb2hqh.ent.gz pdb2hqh.ent.gz | 73.8 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  2hqh.json.gz 2hqh.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| 文書・要旨 |  2hqh_validation.pdf.gz 2hqh_validation.pdf.gz | 462.3 KB | 表示 |  wwPDB検証レポート wwPDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  2hqh_full_validation.pdf.gz 2hqh_full_validation.pdf.gz | 466.3 KB | 表示 | |
| XML形式データ |  2hqh_validation.xml.gz 2hqh_validation.xml.gz | 21.4 KB | 表示 | |
| CIF形式データ |  2hqh_validation.cif.gz 2hqh_validation.cif.gz | 32.1 KB | 表示 | |
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/hq/2hqh https://data.pdbj.org/pub/pdb/validation_reports/hq/2hqh ftp://data.pdbj.org/pub/pdb/validation_reports/hq/2hqh ftp://data.pdbj.org/pub/pdb/validation_reports/hq/2hqh | HTTPS FTP |
-関連構造データ
| 関連構造データ | |
|---|---|
| 類似構造データ |
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| 5 | 
| ||||||||
| 単位格子 |
| ||||||||
| Components on special symmetry positions |
| ||||||||
| 詳細 | The biological assembly is a dimer of chain A and E, or B and F, or C and G, or D and H |
- 要素
要素
| #1: タンパク質 | 分子量: 9928.076 Da / 分子数: 4 / 断片: CAP-Gly domain, residues 15-107 / 由来タイプ: 組換発現 / 由来: (組換発現)  Homo sapiens (ヒト) / 遺伝子: DCTN1 / プラスミド: pET15b / 生物種 (発現宿主): Escherichia coli / 発現宿主: Homo sapiens (ヒト) / 遺伝子: DCTN1 / プラスミド: pET15b / 生物種 (発現宿主): Escherichia coli / 発現宿主:  #2: タンパク質・ペプチド | 分子量: 2928.173 Da / 分子数: 4 / 断片: second zinc finger domain, residues 1405-1427 / 由来タイプ: 組換発現 / 由来: (組換発現)  Homo sapiens (ヒト) / 遺伝子: RSN, CYLN1 / プラスミド: pGEX-4T1 / 生物種 (発現宿主): Escherichia coli / 発現宿主: Homo sapiens (ヒト) / 遺伝子: RSN, CYLN1 / プラスミド: pGEX-4T1 / 生物種 (発現宿主): Escherichia coli / 発現宿主:  #3: 化合物 | ChemComp-ZN / #4: 水 | ChemComp-HOH / | |
|---|
-実験情報
-実験
| 実験 | 手法:  X線回折 / 使用した結晶の数: 1 X線回折 / 使用した結晶の数: 1 |
|---|
- 試料調製
試料調製
| 結晶 | マシュー密度: 2.54 Å3/Da / 溶媒含有率: 51.48 % |
|---|---|
| 結晶化 | 温度: 298 K / 手法: 蒸気拡散法, シッティングドロップ法 / pH: 7 詳細: 4M sodium formate, pH 7, VAPOR DIFFUSION, SITTING DROP, temperature 298K |
-データ収集
| 回折 | 平均測定温度: 100 K | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 放射光源 | 由来:  シンクロトロン / サイト: シンクロトロン / サイト:  APS APS  / ビームライン: 19-BM / 波長: 0.9791, 1.2826, 1.2830, 1.2694 / ビームライン: 19-BM / 波長: 0.9791, 1.2826, 1.2830, 1.2694 | |||||||||||||||
| 検出器 | タイプ: CUSTOM-MADE / 検出器: CCD / 日付: 2005年6月5日 | |||||||||||||||
| 放射 | プロトコル: MAD / 単色(M)・ラウエ(L): M / 散乱光タイプ: x-ray | |||||||||||||||
| 放射波長 |
| |||||||||||||||
| 反射 | 解像度: 1.8→45.7 Å / Num. all: 48867 / Num. obs: 49011 / % possible obs: 100 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Biso Wilson estimate: 14 Å2 | |||||||||||||||
| 反射 シェル | 解像度: 1.8→1.86 Å / % possible all: 100 |
- 解析
解析
| ソフトウェア |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 精密化 | 構造決定の手法:  多波長異常分散 / 解像度: 1.8→45.68 Å / σ(F): 0 / σ(I): 0 / 立体化学のターゲット値: Engh & Huber 多波長異常分散 / 解像度: 1.8→45.68 Å / σ(F): 0 / σ(I): 0 / 立体化学のターゲット値: Engh & Huber
| |||||||||||||||||||||||||
| 原子変位パラメータ | Biso mean: 22.6 Å2
| |||||||||||||||||||||||||
| 精密化ステップ | サイクル: LAST / 解像度: 1.8→45.68 Å
|
 ムービー
ムービー コントローラー
コントローラー












 PDBj
PDBj























