[English] 日本語
 Yorodumi
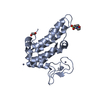
Yorodumi- PDB-2hhi: The solution structure of antigen MPT64 from Mycobacterium tuberc... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2hhi | ||||||
|---|---|---|---|---|---|---|---|
| Title | The solution structure of antigen MPT64 from Mycobacterium tuberculosis defines a novel class of beta-grasp proteins | ||||||
 Components Components | Immunogenic protein MPT64 | ||||||
 Keywords Keywords | UNKNOWN FUNCTION / MPT64 / Secreted Antigen / Tuberculosis / Residual Dipolar Coupling / NMR Solution Structure / beta-grasp | ||||||
| Function / homology |  Function and homology information Function and homology informationcell wall / zymogen binding / peptidoglycan-based cell wall / cellular response to starvation / extracellular region Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method | SOLUTION NMR / Torsion Angle Dynamics, RDC refinement using CNS 1.1 | ||||||
 Authors Authors | Wang, Z. / Potter, B.M. / Gray, A.M. / Sacksteder, K.A. / Geisbrecht, B.V. / Laity, J.H. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2007 Journal: J.Mol.Biol. / Year: 2007Title: The Solution Structure of Antigen MPT64 from Mycobacterium tuberculosis Defines a New Family of Beta-Grasp Proteins. Authors: Wang, Z. / Potter, B.M. / Gray, A.M. / Sacksteder, K.A. / Geisbrecht, B.V. / Laity, J.H. #1:  Journal: Protein Expr.Purif. / Year: 2006 Journal: Protein Expr.Purif. / Year: 2006Title: Design and optimization of a recombinant system for large-scale production of the MPT64 antigen from Mycobacterium tuberculosis. Authors: Geisbrecht, B.V. / Nikonenko, B. / Samala, R. / Nakamura, R. / Nacy, C.A. / Sacksteder, K.A. #2:  Journal: J.Biomol.NMR / Year: 2005 Journal: J.Biomol.NMR / Year: 2005Title: NMR assignment of protein Rv1980c from Mycobacterium tuberculosis Authors: Danahy, J.M. / Potter, B.M. / Geisbrecht, B.V. / Laity, J.H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2hhi.cif.gz 2hhi.cif.gz | 1.4 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2hhi.ent.gz pdb2hhi.ent.gz | 1.2 MB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2hhi.json.gz 2hhi.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hh/2hhi https://data.pdbj.org/pub/pdb/validation_reports/hh/2hhi ftp://data.pdbj.org/pub/pdb/validation_reports/hh/2hhi ftp://data.pdbj.org/pub/pdb/validation_reports/hh/2hhi | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 22375.855 Da / Num. of mol.: 1 / Fragment: Recombinant form of mature polypeptide Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample conditions | Ionic strength: no salt / pH: 6.9 / Temperature: 303 K |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Varian INOVA / Manufacturer: Varian / Model: INOVA / Field strength: 600 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: Torsion Angle Dynamics, RDC refinement using CNS 1.1 Software ordinal: 1 Details: The initial structures were calculated through TAD protocol of CNS with 3761 NOE-derived distance contraints and 292 dihedral angle restraints. The final structures were refined using JNH ...Details: The initial structures were calculated through TAD protocol of CNS with 3761 NOE-derived distance contraints and 292 dihedral angle restraints. The final structures were refined using JNH and JCaCO Residual Dipolar Coupling (RDC) in Cartesian space with same NOE, dihedral angle restraints and 139 JNH RDCs and 114 JCaCO RDCs from stretched gel. | ||||||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 50 / Conformers submitted total number: 24 |
 Movie
Movie Controller
Controller











 PDBj
PDBj HNCA
HNCA