| Entry | Database: PDB / ID: 2ckk
|
|---|
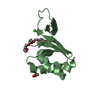
| Title | High resolution crystal structure of the human kin17 C-terminal domain containing a kow motif |
|---|
 Components Components | KIN17 |
|---|
 Keywords Keywords | NUCLEAR PROTEIN / BETA BARREL / RIBOSOMAL PROTEIN / RIBONUCLEOPROTEIN |
|---|
| Function / homology |  Function and homology information Function and homology information
Protein methylation / nuclear matrix / mRNA processing / double-stranded DNA binding / DNA recombination / DNA replication / DNA repair / DNA damage response / protein-containing complex / DNA binding ...Protein methylation / nuclear matrix / mRNA processing / double-stranded DNA binding / DNA recombination / DNA replication / DNA repair / DNA damage response / protein-containing complex / DNA binding / RNA binding / zinc ion binding / nucleoplasm / nucleus / cytosolSimilarity search - Function DNA/RNA-binding protein Kin17, WH-like domain / KIN17-like protein / DNA/RNA-binding protein KIN17, WH-like domain superfamily / KN17, SH3-like C-terminal domain / Kin17, KOW domain / : / KIN17 WH-like domain / KN17 SH3-like domain / KIN17 C-terminal SH3 domain / KIN17 C2H2-type zinc finger domain ...DNA/RNA-binding protein Kin17, WH-like domain / KIN17-like protein / DNA/RNA-binding protein KIN17, WH-like domain superfamily / KN17, SH3-like C-terminal domain / Kin17, KOW domain / : / KIN17 WH-like domain / KN17 SH3-like domain / KIN17 C-terminal SH3 domain / KIN17 C2H2-type zinc finger domain / Domain of Kin17 curved DNA-binding protein / SH3 type barrels. - #30 / SH3 type barrels. - #140 / Zinc finger C2H2 superfamily / Zinc finger C2H2 type domain signature. / SH3 type barrels. / Ribosomal protein L2, domain 2 / Roll / Mainly BetaSimilarity search - Domain/homology |
|---|
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SIRAS / Resolution: 1.45 Å SIRAS / Resolution: 1.45 Å |
|---|
 Authors Authors | le Maire, A. / Schiltz, M. / Pinon-Lataillade, G. / Stura, E. / Couprie, J. / Gondry, M. / Angulo-Mora, J. / Zinn-Justin, S. |
|---|
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2006 Journal: J.Mol.Biol. / Year: 2006
Title: A Tandem of SH3-Like Domains Participates in RNA Binding in Kin17, a Human Protein Activated in Response to Genotoxics.
Authors: Le Maire, A. / Schiltz, M. / Stura, E.A. / Pinon-Lataillade, G. / Couprie, J. / Moutiez, M. / Gondry, M. / Angulo, J.F. / Zinn-Justin, S. |
|---|
| History | | Deposition | Apr 20, 2006 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Oct 4, 2006 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jan 30, 2019 | Group: Data collection / Experimental preparation / Category: exptl_crystal_grow / Item: _exptl_crystal_grow.method |
|---|
| Revision 1.2 | Feb 6, 2019 | Group: Data collection / Experimental preparation / Category: exptl_crystal_grow / Item: _exptl_crystal_grow.temp |
|---|
| Revision 1.3 | May 8, 2024 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Other
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_database_status / struct_site
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_database_status.status_code_sf / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information HOMO SAPIENS (human)
HOMO SAPIENS (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  SIRAS / Resolution: 1.45 Å
SIRAS / Resolution: 1.45 Å  Authors
Authors Citation
Citation Journal: J.Mol.Biol. / Year: 2006
Journal: J.Mol.Biol. / Year: 2006 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2ckk.cif.gz
2ckk.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2ckk.ent.gz
pdb2ckk.ent.gz PDB format
PDB format 2ckk.json.gz
2ckk.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/ck/2ckk
https://data.pdbj.org/pub/pdb/validation_reports/ck/2ckk ftp://data.pdbj.org/pub/pdb/validation_reports/ck/2ckk
ftp://data.pdbj.org/pub/pdb/validation_reports/ck/2ckk Links
Links Assembly
Assembly
 Components
Components HOMO SAPIENS (human) / Production host:
HOMO SAPIENS (human) / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ESRF
ESRF  / Beamline: BM30A / Wavelength: 1.13
/ Beamline: BM30A / Wavelength: 1.13  Processing
Processing SIRAS / Resolution: 1.45→32.62 Å / Cor.coef. Fo:Fc: 0.975 / Cor.coef. Fo:Fc free: 0.958 / SU B: 2.035 / SU ML: 0.036 / Cross valid method: THROUGHOUT / σ(F): 2 / ESU R: 0.064 / ESU R Free: 0.06 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS.
SIRAS / Resolution: 1.45→32.62 Å / Cor.coef. Fo:Fc: 0.975 / Cor.coef. Fo:Fc free: 0.958 / SU B: 2.035 / SU ML: 0.036 / Cross valid method: THROUGHOUT / σ(F): 2 / ESU R: 0.064 / ESU R Free: 0.06 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. Movie
Movie Controller
Controller











 PDBj
PDBj











