[English] 日本語
 Yorodumi
Yorodumi- PDB-1yzd: Crystal structure of an RNA duplex containing a site specific 2'-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1yzd | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of an RNA duplex containing a site specific 2'-amine substitution at a C-G Watson-Crick base pair | ||||||||||||||||||
 Components Components | RNA 5'-R(* Keywords KeywordsRNA / 2'-AMINE / DUPLEX | Function / homology | RNA / RNA (> 10) |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.35 Å MOLECULAR REPLACEMENT / Resolution: 2.35 Å  Authors AuthorsGherghe, C.M. / Krahn, J.M. / Weeks, K.M. |  Citation Citation Journal: J.Am.Chem.Soc. / Year: 2005 Journal: J.Am.Chem.Soc. / Year: 2005Title: Crystal structures, reactivity and inferred acylation transition States for 2'-amine substituted RNA. Authors: Gherghe, C.M. / Krahn, J.M. / Weeks, K.M. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1yzd.cif.gz 1yzd.cif.gz | 37.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1yzd.ent.gz pdb1yzd.ent.gz | 25.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1yzd.json.gz 1yzd.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yz/1yzd https://data.pdbj.org/pub/pdb/validation_reports/yz/1yzd ftp://data.pdbj.org/pub/pdb/validation_reports/yz/1yzd ftp://data.pdbj.org/pub/pdb/validation_reports/yz/1yzd | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 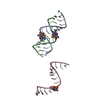
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 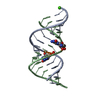
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: RNA chain | Mass: 5097.094 Da / Num. of mol.: 3 / Source method: obtained synthetically / Details: chemically synthesized #2: Chemical | ChemComp-CA / | #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.11 Å3/Da / Density % sol: 41.76 % | ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 273 K / Method: vapor diffusion, sitting drop / pH: 8 Details: MPD, sodium cacodylate, spermidine, magnesium chloride, calcium chloride, pH 8.0, VAPOR DIFFUSION, SITTING DROP, temperature 273K | ||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU / Wavelength: 1.5418 |
| Detector | Type: RIGAKU RAXIS IV++ / Detector: IMAGE PLATE / Date: Feb 24, 2004 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.18→31.7 Å / Num. all: 7277 / Num. obs: 6646 / % possible obs: 91.4 % / Observed criterion σ(I): 1 / Redundancy: 21.79 % / Biso Wilson estimate: 28.7 Å2 / Rmerge(I) obs: 0.077 / Rsym value: 0.06 / Net I/σ(I): 15.2 |
| Reflection shell | Resolution: 2.18→2.28 Å / Rmerge(I) obs: 0.387 / Mean I/σ(I) obs: 3.37 / Num. unique all: 405 / Rsym value: 0.37 / % possible all: 57.4 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.35→8.07 Å / Rfactor Rfree error: 0.017 / Data cutoff high absF: 852998.37 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 MOLECULAR REPLACEMENT / Resolution: 2.35→8.07 Å / Rfactor Rfree error: 0.017 / Data cutoff high absF: 852998.37 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 32.0361 Å2 / ksol: 0.405429 e/Å3 | ||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 30.5 Å2
| ||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.35→8.07 Å
| ||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.35→2.49 Å / Rfactor Rfree error: 0.067 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller





 PDBj
PDBj
































