[English] 日本語
 Yorodumi
Yorodumi- PDB-2tpk: AN INVESTIGATION OF THE STRUCTURE OF THE PSEUDOKNOT WITHIN THE GE... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2tpk | ||||||
|---|---|---|---|---|---|---|---|
| Title | AN INVESTIGATION OF THE STRUCTURE OF THE PSEUDOKNOT WITHIN THE GENE 32 MESSENGER RNA OF BACTERIOPHAGE T2 USING HETERONUCLEAR NMR METHODS | ||||||
 Components Components | RNA (MRNA PSEUDOKNOT) | ||||||
 Keywords Keywords | RNA / PSEUDOKNOT / T2 / RIBONUCLEIC ACID | ||||||
| Function / homology | RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / DISTANCE GEOMETRY-SIMULATED ANNEALING, ENERGY MINIZATION PROTOCOLS | ||||||
 Authors Authors | Holland, J.A. / Hansen, M.R. / Du, Z. / Hoffman, D.W. | ||||||
 Citation Citation |  Journal: Rna / Year: 1999 Journal: Rna / Year: 1999Title: An examination of coaxial stacking of helical stems in a pseudoknot motif: the gene 32 messenger RNA pseudoknot of bacteriophage T2. Authors: Holland, J.A. / Hansen, M.R. / Du, Z. / Hoffman, D.W. #1:  Journal: Nucleic Acids Res. / Year: 1997 Journal: Nucleic Acids Res. / Year: 1997Title: An NMR and Mutational Study of the Pseudoknot within the Gene 32 Mrna of Bacteriophage T2: Insights Into a Family of Structurally Related RNA Pseudoknots Authors: Du, Z. / Hoffman, D.W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2tpk.cif.gz 2tpk.cif.gz | 31.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2tpk.ent.gz pdb2tpk.ent.gz | 21.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2tpk.json.gz 2tpk.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/tp/2tpk https://data.pdbj.org/pub/pdb/validation_reports/tp/2tpk ftp://data.pdbj.org/pub/pdb/validation_reports/tp/2tpk ftp://data.pdbj.org/pub/pdb/validation_reports/tp/2tpk | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 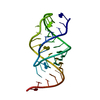
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 11509.891 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: RNA WAS CHEMICALLY SYNTHESIZED FROM GENE 32 OF BACTERIOPHAGE T2. |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||||||||||
| NMR details | Text: THE STRUCTURE WAS DETERMINED USING NMR SPECTROSCOPY ON 13C,15N-LABELED AND SELECTIVELY DEUTERATED T2 PSEUDOKNOT SAMPLE |
- Sample preparation
Sample preparation
| Sample conditions | pH: 6.5 / Temperature: 293 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Manufacturer: Varian / Field strength: 500 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: DISTANCE GEOMETRY-SIMULATED ANNEALING, ENERGY MINIZATION PROTOCOLS Software ordinal: 1 Details: REFINEMNET DETAILS CAN BE FOUND IN THE JRNL CITATION ABOVE. | ||||||||||||||||
| NMR ensemble | Conformers calculated total number: 19 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller











 PDBj
PDBj





























 HSQC
HSQC