[English] 日本語
 Yorodumi
Yorodumi- PDB-1yn2: Solution structure of the Neurospora VS ribozyme stem-loop V in t... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1yn2 | ||||||
|---|---|---|---|---|---|---|---|
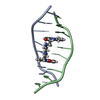
| Title | Solution structure of the Neurospora VS ribozyme stem-loop V in the presence of MgCl2 with modeling of bound manganese ions | ||||||
 Components Components | VS RIBOZYME STEM-LOOP V | ||||||
 Keywords Keywords | RNA / U-turn / hairpin / magnesium ions / manganese ions / paramagnetic | ||||||
| Function / homology | : / RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / SIMULATED ANNEALING, MOLECULAR DYNAMICS | ||||||
| Model type details | minimized average | ||||||
 Authors Authors | Campbell, D.O. / Legault, P. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2006 Journal: Biochemistry / Year: 2006Title: NMR structure of varkud satellite ribozyme stem-loop v in the presence of magnesium ions and localization of metal-binding sites Authors: Campbell, D.O. / Bouchard, P. / Desjardins, G. / Legault, P. #1:  Journal: To be Published / Year: 2005 Journal: To be Published / Year: 2005Title: NMR Structure of the VS Ribozyme Stem-Loop V RNA and Magnesium-Ion Binding from Chemical-Shift Mapping Authors: Campbell, D.O. / Legault, P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1yn2.cif.gz 1yn2.cif.gz | 131.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1yn2.ent.gz pdb1yn2.ent.gz | 106.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1yn2.json.gz 1yn2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yn/1yn2 https://data.pdbj.org/pub/pdb/validation_reports/yn/1yn2 ftp://data.pdbj.org/pub/pdb/validation_reports/yn/1yn2 ftp://data.pdbj.org/pub/pdb/validation_reports/yn/1yn2 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 5419.261 Da / Num. of mol.: 1 / Fragment: SL5 / Source method: obtained synthetically Details: THIS SEQUENCE IS DERIVED FROM THE NEUROSPORA VARKUD SATELLITE RIBOZYME | ||
|---|---|---|---|
| #2: Chemical | ChemComp-MN / #3: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||||||
| NMR details | Text: Bound manganese ions were modeled using distance restraints derived from Mn-induced paramagnetic line broadening |
- Sample preparation
Sample preparation
| Details | Contents: 0.8 mM to 2.0 mM of unlabeled, 15N, or 13C/15N SL5 Solvent system: [10 mM d11-Tris pH 7.0, 50 mM NaCl, 0.05 mM NaN3, 40 mM MgCl2] or [ 10 mM d11-Tris pH 7.0, 0.05 mM NaN3, 20 mM or 46 mM MgCl2]; 90% H2O, 10% D2O or 100% D2O; in some cases 0 uM, 10 ...Solvent system: [10 mM d11-Tris pH 7.0, 50 mM NaCl, 0.05 mM NaN3, 40 mM MgCl2] or [ 10 mM d11-Tris pH 7.0, 0.05 mM NaN3, 20 mM or 46 mM MgCl2]; 90% H2O, 10% D2O or 100% D2O; in some cases 0 uM, 10 uM, 20 uM, 40 uM, or 80 uM MnCl2 were added. |
|---|---|
| Sample conditions | Ionic strength: variable (see sample details) / pH: 7 / Pressure: ambient / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Varian INOVA / Manufacturer: Varian / Model: INOVA / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: SIMULATED ANNEALING, MOLECULAR DYNAMICS / Software ordinal: 1 Details: Conformer 1 represents the minimized average structure, the 10 following structures represent the ensemble of low-energy structures | ||||||||||||||||||||||||
| NMR representative | Selection criteria: minimized average structure | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 46 / Conformers submitted total number: 11 |
 Movie
Movie Controller
Controller







 PDBj
PDBj
































 HSQC
HSQC