+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 1xox | ||||||
|---|---|---|---|---|---|---|---|
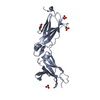
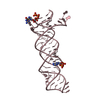
| タイトル | SOLUTION STRUCTURE OF HUMAN SURVIVIN | ||||||
 要素 要素 | Apoptosis inhibitor survivin | ||||||
 キーワード キーワード | APOPTOSIS / Bir Domain | ||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報survivin complex / establishment of chromosome localization / positive regulation of mitotic sister chromatid separation / positive regulation of mitotic cytokinesis / positive regulation of exit from mitosis / positive regulation of mitotic cell cycle spindle assembly checkpoint / mitotic spindle midzone assembly / positive regulation of attachment of mitotic spindle microtubules to kinetochore / interphase microtubule organizing center / chromosome passenger complex ...survivin complex / establishment of chromosome localization / positive regulation of mitotic sister chromatid separation / positive regulation of mitotic cytokinesis / positive regulation of exit from mitosis / positive regulation of mitotic cell cycle spindle assembly checkpoint / mitotic spindle midzone assembly / positive regulation of attachment of mitotic spindle microtubules to kinetochore / interphase microtubule organizing center / chromosome passenger complex / protein-containing complex localization / cobalt ion binding / cysteine-type endopeptidase inhibitor activity / nuclear chromosome / mitotic spindle assembly checkpoint signaling / TP53 regulates transcription of several additional cell death genes whose specific roles in p53-dependent apoptosis remain uncertain / cytoplasmic microtubule / cysteine-type endopeptidase inhibitor activity involved in apoptotic process / mitotic cytokinesis / SUMOylation of DNA replication proteins / chromosome, centromeric region / mitotic spindle assembly / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / Resolution of Sister Chromatid Cohesion / centriole / tubulin binding / positive regulation of mitotic cell cycle / : / spindle microtubule / RHO GTPases Activate Formins / sensory perception of sound / kinetochore / small GTPase binding / spindle / Separation of Sister Chromatids / G2/M transition of mitotic cell cycle / microtubule cytoskeleton / mitotic cell cycle / protein-folding chaperone binding / Neddylation / midbody / microtubule binding / Interleukin-4 and Interleukin-13 signaling / microtubule / protein heterodimerization activity / protein phosphorylation / cell division / negative regulation of DNA-templated transcription / positive regulation of cell population proliferation / negative regulation of apoptotic process / apoptotic process / enzyme binding / protein homodimerization activity / protein-containing complex / zinc ion binding / nucleoplasm / identical protein binding / nucleus / metal ion binding / cytosol / cytoplasm 類似検索 - 分子機能 | ||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | ||||||
| 手法 | 溶液NMR / simulated annealing | ||||||
| Model type details | minimized average | ||||||
 データ登録者 データ登録者 | Sun, C. / Nettesheim, D. / Liu, Z. / Olejniczak, E.T. | ||||||
 引用 引用 |  ジャーナル: Biochemistry / 年: 2005 ジャーナル: Biochemistry / 年: 2005タイトル: Solution structure of human survivin and its binding interface with smac/diablo 著者: Sun, C. / Nettesheim, D. / Liu, Z. / Olejniczak, E.T. | ||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  1xox.cif.gz 1xox.cif.gz | 91.7 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb1xox.ent.gz pdb1xox.ent.gz | 72.6 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  1xox.json.gz 1xox.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| 文書・要旨 |  1xox_validation.pdf.gz 1xox_validation.pdf.gz | 297.9 KB | 表示 |  wwPDB検証レポート wwPDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  1xox_full_validation.pdf.gz 1xox_full_validation.pdf.gz | 297.6 KB | 表示 | |
| XML形式データ |  1xox_validation.xml.gz 1xox_validation.xml.gz | 5.2 KB | 表示 | |
| CIF形式データ |  1xox_validation.cif.gz 1xox_validation.cif.gz | 7.4 KB | 表示 | |
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/xo/1xox https://data.pdbj.org/pub/pdb/validation_reports/xo/1xox ftp://data.pdbj.org/pub/pdb/validation_reports/xo/1xox ftp://data.pdbj.org/pub/pdb/validation_reports/xo/1xox | HTTPS FTP |
-関連構造データ
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR アンサンブル |
|
- 要素
要素
| #1: タンパク質 | 分子量: 13477.332 Da / 分子数: 2 / 断片: residues 1-117 / 由来タイプ: 組換発現 / 由来: (組換発現)  Homo sapiens (ヒト) / 遺伝子: BIRC5, API4, IAP4 / プラスミド: pet15b / 生物種 (発現宿主): Escherichia coli / 発現宿主: Homo sapiens (ヒト) / 遺伝子: BIRC5, API4, IAP4 / プラスミド: pet15b / 生物種 (発現宿主): Escherichia coli / 発現宿主:  #2: 化合物 | |
|---|
-実験情報
-実験
| 実験 | 手法: 溶液NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR実験 |
| ||||||||||||||||
| NMR実験の詳細 | Text: This structure was determined using standard 3D NMR techniques |
- 試料調製
試料調製
| 試料状態 | イオン強度: 50 mM phosphate / pH: 7.5 / 圧: ambient / 温度: 298 K |
|---|
-NMR測定
| NMRスペクトロメーター |
|
|---|
- 解析
解析
| NMR software | 名称: CNX / バージョン: 2000.1 / 開発者: Brunger / 分類: 精密化 |
|---|---|
| 精密化 | 手法: simulated annealing / ソフトェア番号: 1 詳細: Structures were calculated using 1394 intra-monomer NOE constraints, thirty hydrogen bond restraints derived from an analysis of amide exchange rates and fifty-six torsional restraints from ...詳細: Structures were calculated using 1394 intra-monomer NOE constraints, thirty hydrogen bond restraints derived from an analysis of amide exchange rates and fifty-six torsional restraints from an analysis of the backbone chemical shifts. Dimer structures were calculated using additional inter-monomer NOE constraints. |
| 代表構造 | 選択基準: minimized average structure |
| NMRアンサンブル | 登録したコンフォーマーの数: 1 |
 ムービー
ムービー コントローラー
コントローラー











 PDBj
PDBj














