[English] 日本語
 Yorodumi
Yorodumi- PDB-1xnk: Beta-1,4-xylanase from Chaetomium thermophilum complexed with met... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1xnk | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Beta-1,4-xylanase from Chaetomium thermophilum complexed with methyl thioxylopentoside | ||||||||||||
 Components Components | endoxylanase 11A | ||||||||||||
 Keywords Keywords | HYDROLASE / xylanase / glycoside hydrolase / family 11 / glycosidase / thioinhibitor / sulphur containing inhibitor | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationendo-1,4-beta-xylanase activity / endo-1,4-beta-xylanase / xylan catabolic process Similarity search - Function | ||||||||||||
| Biological species |  Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.55 Å MOLECULAR REPLACEMENT / Resolution: 1.55 Å | ||||||||||||
 Authors Authors | Hakanpaa, J. / Hakulinen, N. / Rouvinen, J. | ||||||||||||
 Citation Citation |  Journal: FEBS J. / Year: 2005 Journal: FEBS J. / Year: 2005Title: Determination of thioxylo-oligosaccharide binding to family 11 xylanases using electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry and X-ray crystallography Authors: Janis, J. / Hakanpaa, J. / Hakulinen, N. / Ibatullin, F.M. / Hoxha, A. / Derrick, P.J. / Rouvinen, J. / Vainiotalo, P. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1xnk.cif.gz 1xnk.cif.gz | 98.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1xnk.ent.gz pdb1xnk.ent.gz | 74.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1xnk.json.gz 1xnk.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xn/1xnk https://data.pdbj.org/pub/pdb/validation_reports/xn/1xnk ftp://data.pdbj.org/pub/pdb/validation_reports/xn/1xnk ftp://data.pdbj.org/pub/pdb/validation_reports/xn/1xnk | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1h1aS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 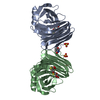
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| |||||||||||||||
| 2 | 
| |||||||||||||||
| 3 | 
| |||||||||||||||
| Unit cell |
| |||||||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 21494.098 Da / Num. of mol.: 2 / Fragment: catalytic domain Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Chaetomium thermophilum (fungus) / Production host: Chaetomium thermophilum (fungus) / Production host:  Hypocrea jecorina (fungus) / References: UniProt: Q8J1V6, endo-1,4-beta-xylanase Hypocrea jecorina (fungus) / References: UniProt: Q8J1V6, endo-1,4-beta-xylanase#2: Polysaccharide | 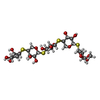  Type: oligosaccharide, Oligosaccharide / Class: Substrate analog / Mass: 476.582 Da / Num. of mol.: 2 Type: oligosaccharide, Oligosaccharide / Class: Substrate analog / Mass: 476.582 Da / Num. of mol.: 2Source method: isolated from a genetically manipulated source Details: oligosaccharide with S-glycosidic bond between monosaccharides References: methyl pentopyranosyl-(1->4)-4-thiopentopyranosyl-(1->4)-4-thio-beta-D-xylopyranosyl-(1->4)-4-thio-beta-D-xylopyranosyl-(1->4)- 4-thio-alpha-D-xylopyranoside #3: Chemical | #4: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.48 Å3/Da / Density % sol: 50.47 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 7 Details: ammonium sulfate, HEPES, pH 7.0, VAPOR DIFFUSION, HANGING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 120 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å |
| Detector | Type: RIGAKU RAXIS IIC / Detector: IMAGE PLATE / Date: Feb 15, 2002 / Details: confocal optics by Osmic |
| Radiation | Monochromator: GRAPHITE / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.55→99 Å / Num. all: 56502 / Num. obs: 56502 / % possible obs: 92.8 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 2.95 % / Biso Wilson estimate: 16.878 Å2 / Rsym value: 0.066 / Net I/σ(I): 11.7 |
| Reflection shell | Resolution: 1.55→1.61 Å / Mean I/σ(I) obs: 1 / Num. unique all: 3124 / Rsym value: 0.296 / % possible all: 52.1 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1H1A Resolution: 1.55→99 Å / Isotropic thermal model: isotropic / σ(F): 0 / Stereochemistry target values: Engh & Huber
| |||||||||||||||||||||||||
| Displacement parameters | Biso mean: 17.184 Å2 | |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.55→99 Å
| |||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj



