+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1w3m | ||||||
|---|---|---|---|---|---|---|---|
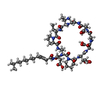
| Title | Crystal structure of tsushimycin | ||||||
 Components Components | TSUSHIMYCIN | ||||||
 Keywords Keywords | ANTIBIOTIC / AMPHOMYCIN / DAPTOMYCIN / LIPOPETIDE / ANTIBACTERIAL | ||||||
| Function / homology | Tsushimycin / ETHANOL / Delta-3isotetradecenoic acid / :  Function and homology information Function and homology information | ||||||
| Biological species |  ACTINOPLANES FRIULIENSIS (bacteria) ACTINOPLANES FRIULIENSIS (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / DIRECT METHODS / Resolution: 1 Å SYNCHROTRON / DIRECT METHODS / Resolution: 1 Å | ||||||
 Authors Authors | Bunkoczi, G. / Vertesy, L. / Sheldrick, G.M. | ||||||
 Citation Citation |  Journal: Acta Crystallogr. D Biol. Crystallogr. / Year: 2005 Journal: Acta Crystallogr. D Biol. Crystallogr. / Year: 2005Title: Structure of the lipopeptide antibiotic tsushimycin. Authors: Bunkoczi, G. / Vertesy, L. / Sheldrick, G.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1w3m.cif.gz 1w3m.cif.gz | 91.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1w3m.ent.gz pdb1w3m.ent.gz | 71 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1w3m.json.gz 1w3m.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1w3m_validation.pdf.gz 1w3m_validation.pdf.gz | 525.7 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1w3m_full_validation.pdf.gz 1w3m_full_validation.pdf.gz | 539.1 KB | Display | |
| Data in XML |  1w3m_validation.xml.gz 1w3m_validation.xml.gz | 16.1 KB | Display | |
| Data in CIF |  1w3m_validation.cif.gz 1w3m_validation.cif.gz | 20.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/w3/1w3m https://data.pdbj.org/pub/pdb/validation_reports/w3/1w3m ftp://data.pdbj.org/pub/pdb/validation_reports/w3/1w3m ftp://data.pdbj.org/pub/pdb/validation_reports/w3/1w3m | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 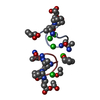
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 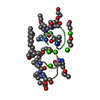
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | 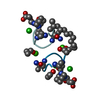
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
-Protein/peptide , 1 types, 12 molecules ABCDEFGHIJKL
-Non-polymers , 5 types, 255 molecules ABCDEFGHIJKL









| #2: Chemical | ChemComp-LNG /   Type: Lipopeptide / Class: Antibiotic / Mass: 226.355 Da / Num. of mol.: 12 / Source method: obtained synthetically / Formula: C14H26O2 Type: Lipopeptide / Class: Antibiotic / Mass: 226.355 Da / Num. of mol.: 12 / Source method: obtained synthetically / Formula: C14H26O2Details: TSUSHIMYCIN IS A CYCLIC LIPOPEPTIDE. THE SCAFFOLD IS MADE OF TWO PARTS: (1) ASP1 N-TERM EXOCYCLIC PART (2) A DECAPEPTIDE RING. RESIDUE 2 AND 11 FORM A PEPTIDE BOND. A TETRADECENOIC FATTY ...Details: TSUSHIMYCIN IS A CYCLIC LIPOPEPTIDE. THE SCAFFOLD IS MADE OF TWO PARTS: (1) ASP1 N-TERM EXOCYCLIC PART (2) A DECAPEPTIDE RING. RESIDUE 2 AND 11 FORM A PEPTIDE BOND. A TETRADECENOIC FATTY ACID IS LINKED TO THE RESIDUE 1 References: Tsushimycin #3: Chemical | ChemComp-CA / #4: Chemical | ChemComp-CL / #5: Chemical | ChemComp-EOH / #6: Water | ChemComp-HOH / | |
|---|
-Details
| Compound details | TSUSHIMYCIN IS A CYCLIC DODEDECAMER LIPOPETIDE. HERE, TSUSHIMYCIN IS REPRESENTED BY GROUPING ...TSUSHIMYCI |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.41 Å3/Da / Density % sol: 40 % / Description: NONE |
|---|---|
| Crystal grow | Method: vapor diffusion, hanging drop / pH: 4 Details: 0.1 M NAAC/HAC PH=4.0 0.12 M HAC 38% ETOH 0.80 M CACL2 0.40 M 1,6-HEXANEDIOL |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  BESSY BESSY  / Beamline: 14.1 / Wavelength: 0.9 / Beamline: 14.1 / Wavelength: 0.9 |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Dec 12, 2003 / Details: MIRRORS |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9 Å / Relative weight: 1 |
| Reflection | Resolution: 1→26.02 Å / Num. obs: 355145 / % possible obs: 96.2 % / Redundancy: 4.5 % / Rmerge(I) obs: 0.1 / Net I/σ(I): 9.49 |
| Reflection shell | Resolution: 1→1.1 Å / Redundancy: 3.62 % / Rmerge(I) obs: 0.5 / Mean I/σ(I) obs: 3.35 / % possible all: 92.1 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: DIRECT METHODS / Resolution: 1→99 Å / Num. parameters: 11896 / Num. restraintsaints: 14162 / Cross valid method: FREE R-VALUE / σ(F): 0 Stereochemistry target values: ENGH AND HUBER GENERATED USING CCDC
| |||||||||||||||||||||||||||||||||
| Refine analyze | Num. disordered residues: 26 / Occupancy sum hydrogen: 829 / Occupancy sum non hydrogen: 1259.82 | |||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1→99 Å
| |||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj