[English] 日本語
 Yorodumi
Yorodumi- PDB-1tcj: STRUCTURE-ACTIVITY RELATIONSHIPS OF MU-CONOTOXIN GIIIA: STRUCTURE... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1tcj | ||||||
|---|---|---|---|---|---|---|---|
| Title | STRUCTURE-ACTIVITY RELATIONSHIPS OF MU-CONOTOXIN GIIIA: STRUCTURE DETERMINATION OF ACTIVE AND INACTIVE SODIUM CHANNEL BLOCKER PEPTIDES BY NMR AND SIMULATED ANNEALING CALCULATIONS | ||||||
 Components Components | MU-CONOTOXIN GIIIA | ||||||
 Keywords Keywords | NEUROTOXIN | ||||||
| Function / homology | Mu-Conotoxin / Conotoxin, mu-type / Mu-conotoxin family signature. / sodium channel inhibitor activity / toxin activity / extracellular region / Mu-conotoxin GIIIA Function and homology information Function and homology information | ||||||
| Biological species |  Conus geographus (geography cone) Conus geographus (geography cone) | ||||||
| Method | SOLUTION NMR | ||||||
 Authors Authors | Kohda, D. / Lancelin, J.-M. / Inagaki, F. / Wakamatsu, K. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1992 Journal: Biochemistry / Year: 1992Title: Structure-activity relationships of mu-conotoxin GIIIA: structure determination of active and inactive sodium channel blocker peptides by NMR and simulated annealing calculations. Authors: Wakamatsu, K. / Kohda, D. / Hatanaka, H. / Lancelin, J.M. / Ishida, Y. / Oya, M. / Nakamura, H. / Inagaki, F. / Sato, K. #1:  Journal: Biochemistry / Year: 1991 Journal: Biochemistry / Year: 1991Title: Tertiary Structure of Conotoxin Giiia in Aqueous Solution Authors: Lancelin, J.-M. / Kohda, D. / Tate, S.-I. / Yanagawa, Y. / Abe, T. / Satake, M. / Inagaki, F. #2:  Journal: J.Biol.Chem. / Year: 1991 Journal: J.Biol.Chem. / Year: 1991Title: Active Site of Mu-Conotoxin Giiia, a Peptide Blocker of Muscle Sodium Channels Authors: Sato, K. / Ishida, Y. / Wakamatsu, K. / Kato, R. / Honda, H. / Ohizumi, Y. / Nakamura, H. / Ohya, M. / Lancelin, J.-M. / Kohda, D. / Inagaki, F. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1tcj.cif.gz 1tcj.cif.gz | 77.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1tcj.ent.gz pdb1tcj.ent.gz | 57.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1tcj.json.gz 1tcj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/tc/1tcj https://data.pdbj.org/pub/pdb/validation_reports/tc/1tcj ftp://data.pdbj.org/pub/pdb/validation_reports/tc/1tcj ftp://data.pdbj.org/pub/pdb/validation_reports/tc/1tcj | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 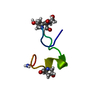
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Atom site foot note | 1: RESIDUE HYP 7 IS A CIS HYDROXYPROLINE. / 2: THE C-TERMINAL CARBOXYLATE IS AMIDATED. | |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 2621.139 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Conus geographus (geography cone) / References: UniProt: P01523 Conus geographus (geography cone) / References: UniProt: P01523 |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Processing
Processing
| NMR software | Name:  X-PLOR / Version: 2.1 / Developer: BRUNGER / Classification: refinement X-PLOR / Version: 2.1 / Developer: BRUNGER / Classification: refinement |
|---|---|
| NMR ensemble | Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller








 PDBj
PDBj