[English] 日本語
 Yorodumi
Yorodumi- PDB-1t9e: NMR solution structure of a disulfide analogue of the cyclic sunf... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1t9e | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR solution structure of a disulfide analogue of the cyclic sunflower trypsin inhibitor SFTI-1 | ||||||
 Components Components | Trypsin inhibitor 1 | ||||||
 Keywords Keywords | HYDROLASE/HYDROLASE INHIBITOR / sunflower trypsin inhibitor / Disulfide mutant / HYDROLASE-HYDROLASE INHIBITOR COMPLEX | ||||||
| Function / homology | negative regulation of endopeptidase activity / endopeptidase inhibitor activity / serine-type endopeptidase inhibitor activity / protease binding / Trypsin inhibitor 1 Function and homology information Function and homology information | ||||||
| Biological species |  | ||||||
| Method | SOLUTION NMR / Structures were calculated using torsion angle dynamics, refined using cartesian dynamics, energy minimization in solvent using CNS. | ||||||
 Authors Authors | Korsinczky, M.L.J. / Clark, R.J. / Craik, D.J. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2005 Journal: Biochemistry / Year: 2005Title: Disulfide bond mutagenesis and the structure and function of the head-to-tail macrocyclic trypsin inhibitor SFTI-1 Authors: Korsinczky, M.L.J. / Clark, R.J. / Craik, D.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1t9e.cif.gz 1t9e.cif.gz | 78.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1t9e.ent.gz pdb1t9e.ent.gz | 53.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1t9e.json.gz 1t9e.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/t9/1t9e https://data.pdbj.org/pub/pdb/validation_reports/t9/1t9e ftp://data.pdbj.org/pub/pdb/validation_reports/t9/1t9e ftp://data.pdbj.org/pub/pdb/validation_reports/t9/1t9e | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 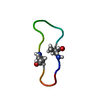
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 1499.753 Da / Num. of mol.: 1 / Mutation: C3(ABA), C11(ABA) / Source method: obtained synthetically Details: The peptide was synthesized using standard solid phase peptide synthesis methods using BOC chemistry. The peptide backbone was cyclicized in solution by addition of HBTU and DIEA which ...Details: The peptide was synthesized using standard solid phase peptide synthesis methods using BOC chemistry. The peptide backbone was cyclicized in solution by addition of HBTU and DIEA which resulted in the formation of a peptide bond between D14 and G1. The sequence of the peptide is naturally found in Helianthus Annus (sunflower) Source: (synth.)  |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||
| NMR details | Text: This structure was determined using standard 2D homonuclear techniques. |
- Sample preparation
Sample preparation
| Details |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions | pH: 4.5 / Pressure: ambient / Temperature: 290 K |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Bruker DMX / Manufacturer: Bruker / Model: DMX / Field strength: 750 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: Structures were calculated using torsion angle dynamics, refined using cartesian dynamics, energy minimization in solvent using CNS. Software ordinal: 1 | ||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 50 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller













 PDBj
PDBj