+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1rzs | ||||||
|---|---|---|---|---|---|---|---|
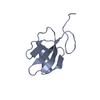
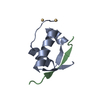
| Title | Solution structure of P22 Cro | ||||||
 Components Components | Regulatory protein cro | ||||||
 Keywords Keywords | TRANSCRIPTION / HELIX-TURN-HELIX / DNA-BINDING PROTEIN / STRUCTURAL EVOLUTION | ||||||
| Function / homology | DNA-binding transcriptional regulator Cro / lambda repressor-like DNA-binding domains / 434 Repressor (Amino-terminal Domain) / Lambda repressor-like, DNA-binding domain superfamily / DNA binding / Orthogonal Bundle / Mainly Alpha / Regulatory protein cro Function and homology information Function and homology information | ||||||
| Biological species |  Enterobacteria phage P22 (virus) Enterobacteria phage P22 (virus) | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
 Authors Authors | Newlove, T. / Konieczka, J.H. / Cordes, M.H. | ||||||
 Citation Citation |  Journal: STRUCTURE / Year: 2004 Journal: STRUCTURE / Year: 2004Title: Secondary structure switching in Cro protein evolution. Authors: Newlove, T. / Konieczka, J.H. / Cordes, M.H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1rzs.cif.gz 1rzs.cif.gz | 393.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1rzs.ent.gz pdb1rzs.ent.gz | 330.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1rzs.json.gz 1rzs.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rz/1rzs https://data.pdbj.org/pub/pdb/validation_reports/rz/1rzs ftp://data.pdbj.org/pub/pdb/validation_reports/rz/1rzs ftp://data.pdbj.org/pub/pdb/validation_reports/rz/1rzs | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 6837.767 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Enterobacteria phage P22 (virus) / Genus: P22-like viruses / Gene: CRO / Plasmid: pET21b / Species (production host): Escherichia coli / Production host: Enterobacteria phage P22 (virus) / Genus: P22-like viruses / Gene: CRO / Plasmid: pET21b / Species (production host): Escherichia coli / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
|
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Bruker DRX / Manufacturer: Bruker / Model: DRX / Field strength: 600 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 Details: 30 structures were calculated using 916 NOE-derived distance restraints, 10 hydrogen bond distance restraints, 41 phi angle restraints and 5 chi1 angle restraints. 23 of 30 calculated ...Details: 30 structures were calculated using 916 NOE-derived distance restraints, 10 hydrogen bond distance restraints, 41 phi angle restraints and 5 chi1 angle restraints. 23 of 30 calculated structures were initially accepted based on no distance restraint violations greater than 0.4 angstroms and no angle restraint violations greater than 5 degrees. Of the 23, 2 were eliminated for having significantly higher energy than the others, leaving 21 structures in the final ensemble. The ordered region of the structure includes residues 1-57. Pairwise RMSDs for the ordered region were 0.53 A (backbone atoms) and 1.29 A (all heavy atoms). None of the backbone angles in the ordered region of any ensemble member fell outside the most favorable and additionally allowed regions of a Ramachandran plot. | ||||||||||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: agreement with distance and angle restraints; two structures were also eliminated due to high energy Conformers calculated total number: 30 / Conformers submitted total number: 21 |
 Movie
Movie Controller
Controller









 PDBj
PDBj


 NMRPipe
NMRPipe