[English] 日本語
 Yorodumi
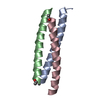
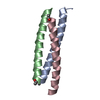
Yorodumi- PDB-1rb6: ANTIPARALLEL TRIMER OF GCN4-LEUCINE ZIPPER CORE MUTANT AS N16A TE... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1rb6 | ||||||
|---|---|---|---|---|---|---|---|
| Title | ANTIPARALLEL TRIMER OF GCN4-LEUCINE ZIPPER CORE MUTANT AS N16A TETRAGONAL FORM | ||||||
 Components Components | General control protein GCN4 | ||||||
 Keywords Keywords | DNA BINDING PROTEIN / COILED COIL / LEUCINE ZIPPER | ||||||
| Function / homology |  Function and homology information Function and homology informationFCERI mediated MAPK activation / protein localization to nuclear periphery / Activation of the AP-1 family of transcription factors / response to amino acid starvation / negative regulation of ribosomal protein gene transcription by RNA polymerase II / positive regulation of cellular response to amino acid starvation / mediator complex binding / Oxidative Stress Induced Senescence / TFIID-class transcription factor complex binding / amino acid biosynthetic process ...FCERI mediated MAPK activation / protein localization to nuclear periphery / Activation of the AP-1 family of transcription factors / response to amino acid starvation / negative regulation of ribosomal protein gene transcription by RNA polymerase II / positive regulation of cellular response to amino acid starvation / mediator complex binding / Oxidative Stress Induced Senescence / TFIID-class transcription factor complex binding / amino acid biosynthetic process / positive regulation of RNA polymerase II transcription preinitiation complex assembly / positive regulation of transcription initiation by RNA polymerase II / cellular response to nutrient levels / cellular response to amino acid starvation / RNA polymerase II transcription regulator complex / DNA-binding transcription activator activity, RNA polymerase II-specific / transcription regulator complex / sequence-specific DNA binding / RNA polymerase II-specific DNA-binding transcription factor binding / DNA-binding transcription factor activity, RNA polymerase II-specific / intracellular signal transduction / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / chromatin binding / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / identical protein binding / nucleus Similarity search - Function | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 1.9 Å MAD / Resolution: 1.9 Å | ||||||
 Authors Authors | Holton, J. / Alber, T. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2004 Journal: Proc.Natl.Acad.Sci.USA / Year: 2004Title: Automated protein crystal structure determination using ELVES. Authors: Holton, J. / Alber, T. #1:  Journal: Nat.Struct.Biol. / Year: 1996 Journal: Nat.Struct.Biol. / Year: 1996Title: An Engineered Allosteric Switch in Leucine-Zipper Oligomerization Authors: Gonzalez Jr., L. / Plecs, J.J. / Alber, T. #2:  Journal: Nature / Year: 1994 Journal: Nature / Year: 1994Title: Crystal Structure of an Isoleucine-Zipper Trimer Authors: Harbury, P.B. / Kim, P.S. / Alber, T. #3:  Journal: Science / Year: 1993 Journal: Science / Year: 1993Title: A Switch between Two-, Three-, and Four-Stranded Coiled Coils in GCN4 Leucine Zipper Mutants Authors: Harbury, P.B. / Zhang, T. / Kim, P.S. / Alber, T. #4:  Journal: Science / Year: 1991 Journal: Science / Year: 1991Title: X-Ray Structure of the GCN4 Leucine Zipper, a two-stranded, parallel coiled coil. Authors: O'Shea, E.K. / Klemm, J.D. / Kim, P.S. / Alber, T. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1rb6.cif.gz 1rb6.cif.gz | 37.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1rb6.ent.gz pdb1rb6.ent.gz | 26.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1rb6.json.gz 1rb6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rb/1rb6 https://data.pdbj.org/pub/pdb/validation_reports/rb/1rb6 ftp://data.pdbj.org/pub/pdb/validation_reports/rb/1rb6 ftp://data.pdbj.org/pub/pdb/validation_reports/rb/1rb6 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein/peptide | Mass: 3988.677 Da / Num. of mol.: 3 / Fragment: LEUCINE-ZIPPER (RESIDUES 249-281) / Mutation: N16A / Source method: obtained synthetically Details: The sequence of the protein is naturally found in Saccharomyces cerevisiae. The protein was chemically synthesized. References: UniProt: P03069 #2: Chemical | ChemComp-CL / #3: Chemical | #4: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.44 Å3/Da / Density % sol: 75 % | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / pH: 7.3 Details: 25 mM phosphate 400 mM NaCl 15% PEG8000, pH 7.3, VAPOR DIFFUSION, SITTING DROP, temperature 298K | |||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS pH: 7 / Method: vapor diffusion | |||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 90 K | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL1-5 / Wavelength: 1.0688, 0.9800, 0.9797, 0.9795, 0.9322 / Beamline: BL1-5 / Wavelength: 1.0688, 0.9800, 0.9797, 0.9795, 0.9322 | ||||||||||||||||||
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Jun 11, 1998 | ||||||||||||||||||
| Radiation | Monochromator: SI(111) / Protocol: MAD / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||
| Radiation wavelength |
| ||||||||||||||||||
| Reflection | Resolution: 1.8→45 Å / Num. all: 20031 / Num. obs: 19898 / % possible obs: 99.6 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 | ||||||||||||||||||
| Reflection shell | Resolution: 1.8→1.9 Å / % possible all: 97.5 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MAD / Resolution: 1.9→15 Å / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber MAD / Resolution: 1.9→15 Å / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→15 Å
| |||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Version: 4 / Classification: refinement | |||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller













 PDBj
PDBj






