[English] 日本語
 Yorodumi
Yorodumi- PDB-1rau: SOLUTION STRUCTURE OF AN UNUSUALLY STABLE RNA TETRAPLEX CONTAININ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1rau | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
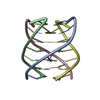
| Title | SOLUTION STRUCTURE OF AN UNUSUALLY STABLE RNA TETRAPLEX CONTAINING G-AND U-QUARTET STRUCTURES | ||||||||||||||||||
 Components Components | RNA (5'-R(* Keywords KeywordsRNA / TETRAPLEX | Function / homology | RNA |  Function and homology information Function and homology informationMethod | SOLUTION NMR |  Authors AuthorsCheong, C. / Moore, P.B. |  Citation Citation Journal: Biochemistry / Year: 1992 Journal: Biochemistry / Year: 1992Title: Solution structure of an unusually stable RNA tetraplex containing G- and U-quartet structures. Authors: Cheong, C. / Moore, P.B. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1rau.cif.gz 1rau.cif.gz | 22 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1rau.ent.gz pdb1rau.ent.gz | 15.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1rau.json.gz 1rau.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ra/1rau https://data.pdbj.org/pub/pdb/validation_reports/ra/1rau ftp://data.pdbj.org/pub/pdb/validation_reports/ra/1rau ftp://data.pdbj.org/pub/pdb/validation_reports/ra/1rau | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 1948.197 Da / Num. of mol.: 4 / Source method: obtained synthetically / Details: CHEMICALLY SYNTHESIZED /  Keywords: RIBONUCLEIC ACID Keywords: RIBONUCLEIC ACID |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR details | Text: ATA COLLECTION PARAMETERS: FREQUENCY: 500 MHZ FOR 1H, 202 MHZ FOR 31P SAMPLE TUBE: 5 MM SPECTRA FOR ASSIGNMENTS: HOMONUCLEAR PROTON 2D, 1H-31P COSY; SPECTRA FOR CONSTRAINTS: NOESY, DQF-COSY, 1H- ...Text: ATA COLLECTION PARAMETERS: FREQUENCY: 500 MHZ FOR 1H, 202 MHZ FOR 31P SAMPLE TUBE: 5 MM SPECTRA FOR ASSIGNMENTS: HOMONUCLEAR PROTON 2D, 1H-31P COSY; SPECTRA FOR CONSTRAINTS: NOESY, DQF-COSY, 1H-31P COSY MOLECULAR CHARACTERISTICS: OLIGOMER STATE: TETRAMERIC MOLECULAR WEIGHT: 13 KD NUCLEOTIDES: 24 SOLUTION CONDITIONS: SOLVENT: H2O AND 100% D2O TEMPERATURES: 5C AND 40C PH: 6.7 AND 5.2 STRAND CONCENTRATION: 2.2 MM ADDITIVES: 10 MM POTASSIUM PHOSPHATE, 50 MM KCL, 0.5 MM EDTA OR 18 MM POTASSIUM PHOSPHATE, 88 MM KCL, 0.9 MM EDTA |
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| |||||||||||||||||||||
| Sample conditions |
|
- Processing
Processing
| Software |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| NMR software | Name:  X-PLOR / Developer: BRUNGER / Classification: refinement X-PLOR / Developer: BRUNGER / Classification: refinement | ||||||||
| Refinement | Software ordinal: 1 Details: STRUCTURE CALCULATION: MOLECULAR DYNAMICS AND ENERGY MINIMIZATION USING RELAXATION MATRIX ROUTINE IN X-PLOR CONSTRAINTS: TOTAL 428 DISTANCE CONSTRAINTS (ABOUT 18/NUCLEOTIDE) 212 FROM ...Details: STRUCTURE CALCULATION: MOLECULAR DYNAMICS AND ENERGY MINIMIZATION USING RELAXATION MATRIX ROUTINE IN X-PLOR CONSTRAINTS: TOTAL 428 DISTANCE CONSTRAINTS (ABOUT 18/NUCLEOTIDE) 212 FROM INTRANUCLEOTIDE NOES 112 FROM INTERNUCLEOTIDE NOES 36 FROM HYDROGEN BONDS 8 FROM EXCHANGEABLE PROTON NOES 60 DERIVED FROM BACKBONE TORSION ANGLES DIHEDRAL ANGLE CONSTRAINTS 104 BACKBONE AND GLYCOSIDIC TORSION ANGLES SUGAR RING TORSION ANGLES CONSTRAINTS TO MAKE QUARTET PLANES PLANAR 4-FOLD ROTATIONAL SYMMETRY CONSTRAINTS QUALITY OF STRUCTURE NUMBER OF REFINED STRUCTURES: 8 RMS DEVIATION: 0.71A FOR ALL HEAVY ATOMS | ||||||||
| NMR constraints | NOE constraints total: 428 / NOE intraresidue total count: 212 / Hydrogen bond constraints total count: 36 | ||||||||
| NMR ensemble | Conformers calculated total number: 8 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller







 PDBj
PDBj