[English] 日本語
 Yorodumi
Yorodumi- PDB-1r1c: PSEUDOMONAS AERUGINOSA W48F/Y72F/H83Q/Y108W-AZURIN RE(PHEN)(CO)3(... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1r1c | ||||||
|---|---|---|---|---|---|---|---|
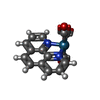
| Title | PSEUDOMONAS AERUGINOSA W48F/Y72F/H83Q/Y108W-AZURIN RE(PHEN)(CO)3(HIS107) | ||||||
 Components Components | Azurin | ||||||
 Keywords Keywords | ELECTRON TRANSPORT / BLUE-COPPER / ELECTRON-TRANSFER / RHENIUM / TUNNELING / RADICAL / EPR | ||||||
| Function / homology |  Function and homology information Function and homology informationtransition metal ion binding / periplasmic space / electron transfer activity / copper ion binding / zinc ion binding / identical protein binding / plasma membrane Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å MOLECULAR REPLACEMENT / Resolution: 1.9 Å | ||||||
 Authors Authors | Miller, J.E. / Gradinaru, C. / Crane, B.R. / Di Bilio, A.J. | ||||||
 Citation Citation |  Journal: J.Am.Chem.Soc. / Year: 2003 Journal: J.Am.Chem.Soc. / Year: 2003Title: Spectroscopy and reactivity of a photogenerated tryptophan radical in a structurally defined protein environment Authors: Miller, J.E. / Gradinaru, C. / Crane, B.R. / Di Bilio, A.J. / Wehbi, W.A. / Un, S. / Winkler, J.R. / Gray, H.B. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1r1c.cif.gz 1r1c.cif.gz | 123.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1r1c.ent.gz pdb1r1c.ent.gz | 96.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1r1c.json.gz 1r1c.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1r1c_validation.pdf.gz 1r1c_validation.pdf.gz | 412.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1r1c_full_validation.pdf.gz 1r1c_full_validation.pdf.gz | 432.3 KB | Display | |
| Data in XML |  1r1c_validation.xml.gz 1r1c_validation.xml.gz | 14.6 KB | Display | |
| Data in CIF |  1r1c_validation.cif.gz 1r1c_validation.cif.gz | 23 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/r1/1r1c https://data.pdbj.org/pub/pdb/validation_reports/r1/1r1c ftp://data.pdbj.org/pub/pdb/validation_reports/r1/1r1c ftp://data.pdbj.org/pub/pdb/validation_reports/r1/1r1c | HTTPS FTP |
-Related structure data
| Related structure data |  1bexS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 13929.798 Da / Num. of mol.: 4 / Mutation: W48F/Y72F/H83Q/Q107H/Y108W Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Chemical | ChemComp-CU1 / #3: Chemical | ChemComp-REP / ( #4: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.95 Å3/Da / Density % sol: 46.59 % | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / pH: 7.5 Details: PEG 4000, lithium nitrate, imidazole, pH 7.5, VAPOR DIFFUSION, SITTING DROP, temperature 298K | ||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  CHESS CHESS  / Beamline: F2 / Wavelength: 0.964 / Wavelength: 0.964 Å / Beamline: F2 / Wavelength: 0.964 / Wavelength: 0.964 Å | |||||||||
| Detector | Type: ADSC QUANTUM 210 / Detector: CCD / Date: Oct 8, 2002 | |||||||||
| Radiation | Monochromator: SI / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||
| Radiation wavelength |
| |||||||||
| Reflection | Resolution: 1.9→40 Å / Num. obs: 76883 / % possible obs: 98.8 % / Redundancy: 3.6 % / Rsym value: 0.075 / Net I/σ(I): 14 | |||||||||
| Reflection shell | Resolution: 1.9→2 Å / Redundancy: 3.61 % / Mean I/σ(I) obs: 3.35 / Rsym value: 0.374 / % possible all: 97.8 | |||||||||
| Reflection | *PLUS Highest resolution: 1.9 Å / Rmerge(I) obs: 0.075 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1BEX Resolution: 1.9→40 Å / Num. parameters: 17719 / Num. restraintsaints: 16833 / Cross valid method: FREE R / σ(F): 0 / Stereochemistry target values: Engh & Huber Details: ANISOTROPIC SCALING APPLIED BY THE METHOD OF PARKIN, MOEZZI & HOPE, J.APPL.CRYST. 28(1995)53-56.
| |||||||||||||||||||||||||||||||||
| Refine analyze | Num. disordered residues: 8 / Occupancy sum hydrogen: 0 / Occupancy sum non hydrogen: 4279 | |||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→40 Å
| |||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.9→2 Å /
| |||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 1.9 Å / % reflection Rfree: 5 % / Rfactor Rfree: 0.26 / Rfactor Rwork: 0.224 | |||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller











 PDBj
PDBj