+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1qtu | ||||||
|---|---|---|---|---|---|---|---|
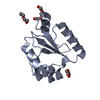
| Title | SOLUTION STRUCTURE OF THE ONCOPROTEIN P13MTCP1 | ||||||
 Components Components | PROTEIN (PRODUCT OF THE MTCP1 ONCOGENE) | ||||||
 Keywords Keywords | GENE REGULATION / BETA BARREL | ||||||
| Function / homology |  Function and homology information Function and homology informationprotein serine/threonine kinase activator activity / intracellular signal transduction / protein kinase binding Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
| Model type details | minimized average | ||||||
 Authors Authors | Guignard, L. / Padilla, A. / Mispelter, J. / Yang, Y.-S. / Stern, M.-H. / Lhoste, J.-M. / Roumestand, C. | ||||||
 Citation Citation |  Journal: J.Biomol.NMR / Year: 2000 Journal: J.Biomol.NMR / Year: 2000Title: Backbone dynamics and solution structure refinement of the 15N-labeled human oncogenic protein p13MTCP1: comparison with X-ray data. Authors: Guignard, L. / Padilla, A. / Mispelter, J. / Yang, Y.S. / Stern, M.H. / Lhoste, J.M. / Roumestand, C. #1:  Journal: J.Biomol.NMR / Year: 1998 Journal: J.Biomol.NMR / Year: 1998Title: Solution structure of the recombinant human oncoprotein p13MTCP1 Authors: Yang, Y.-S. / Guignard, L. / Padilla, A. / Hoh, F. / Strub, M.-P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1qtu.cif.gz 1qtu.cif.gz | 47.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1qtu.ent.gz pdb1qtu.ent.gz | 33.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1qtu.json.gz 1qtu.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qt/1qtu https://data.pdbj.org/pub/pdb/validation_reports/qt/1qtu ftp://data.pdbj.org/pub/pdb/validation_reports/qt/1qtu ftp://data.pdbj.org/pub/pdb/validation_reports/qt/1qtu | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 13680.420 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Cell: T-CELLS / Cell (production host): BL21 / Production host: Homo sapiens (human) / Cell: T-CELLS / Cell (production host): BL21 / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 2mM p13MTCP1 U-15N; 20mM phosphate buffer NA;90% H2O, 10% D2O Solvent system: 90% H2O/10% D2O |
|---|---|
| Sample conditions | pH: 6.5 / Pressure: ambient / Temperature: 303 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software | Name:  Amber / Version: 4.1 / Developer: Kollman. P-A. / Classification: refinement Amber / Version: 4.1 / Developer: Kollman. P-A. / Classification: refinement |
|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 Details: the structures are based on a total of 1773 NOE restraints, 80 dihedral angle restraints |
| NMR representative | Selection criteria: minimized average structure |
| NMR ensemble | Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller














 PDBj
PDBj