[English] 日本語
 Yorodumi
Yorodumi- PDB-1ppk: CRYSTALLOGRAPHIC ANALYSIS OF TRANSITION STATE MIMICS BOUND TO PEN... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ppk | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CRYSTALLOGRAPHIC ANALYSIS OF TRANSITION STATE MIMICS BOUND TO PENICILLOPEPSIN: PHOSPHOROUS-CONTAINING PEPTIDE ANALOGUES | |||||||||
 Components Components | PENICILLOPEPSIN | |||||||||
 Keywords Keywords | HYDROLASE/HYDROLASE INHIBITOR / ACID PROTEINASE / HYDROLASE-HYDROLASE INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpenicillopepsin / aspartic-type endopeptidase activity / proteolysis / extracellular region Similarity search - Function | |||||||||
| Biological species |  Penicillium janthinellum (fungus) Penicillium janthinellum (fungus) | |||||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.8 Å X-RAY DIFFRACTION / Resolution: 1.8 Å | |||||||||
 Authors Authors | Strynadka, N.C.J. / James, M.N.G. | |||||||||
 Citation Citation |  Journal: Biochemistry / Year: 1992 Journal: Biochemistry / Year: 1992Title: Crystallographic analysis of transition-state mimics bound to penicillopepsin: phosphorus-containing peptide analogues. Authors: Fraser, M.E. / Strynadka, N.C. / Bartlett, P.A. / Hanson, J.E. / James, M.N. #1:  Journal: Biochemistry / Year: 1992 Journal: Biochemistry / Year: 1992Title: Crystallographic Analysis of Transition State Mimics Bound to Penicillopepsin: Difluorostatine-and Difluorostatone-Containing Peptides Authors: James, M.N.G. / Sielecki, A.R. / Hayakawa, K. / Gelb, M.H. #2:  Journal: Biological Macromolecules and Assemblies / Year: 1987 Journal: Biological Macromolecules and Assemblies / Year: 1987Title: Aspartic Proteinases and Their Catalytic Pathway Authors: James, M.N.G. / Sielecki, A.R. #3:  Journal: Biochemistry / Year: 1985 Journal: Biochemistry / Year: 1985Title: Stereochemical Analysis of Peptide Bond Hydrolysis Catalyzed by the Aspartic Proteinase Penicillopepsin Authors: James, M.N.G. / Sielecki, A.R. #4:  Journal: Aspartic Proteinases and Their Inhibitors / Year: 1985 Journal: Aspartic Proteinases and Their Inhibitors / Year: 1985Title: X-Ray Diffraction Studies on Penicillopepsin and its Complexes: The Hydrolytic Mechanism Authors: James, M.N.G. / Sielecki, A.R. / Hofmann, T. #5:  Journal: Biochemistry / Year: 1984 Journal: Biochemistry / Year: 1984Title: Effect of Ph on the Activities of Penicillopepsin and Rhizopus Pepsin and a Proposal for the Productive Substrate Binding Mode in Penicillopepsin Authors: Hofmann, T. / Hodges, R.S. / James, M.N.G. #6:  Journal: Peptides: Structure and Function, Proceedings of the of the Eighth American Peptide Symposium Journal: Peptides: Structure and Function, Proceedings of the of the Eighth American Peptide SymposiumYear: 1983 Title: Crystallographic Analysis of a Pepstatin Analogue Binding to the Aspartyl Proteinase Penicillopepsin at 1.8 Angstroms Resolution Authors: James, M.N.G. / Sielecki, A.R. / Moult, J. #7:  Journal: J.Mol.Biol. / Year: 1983 Journal: J.Mol.Biol. / Year: 1983Title: Structure and Refinement of Penicillopepsin at 1.8 Angstroms Resolution Authors: James, M.N.G. / Sielecki, A.R. #8:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1982 Journal: Proc.Natl.Acad.Sci.USA / Year: 1982Title: Conformational Flexibility in the Active Sites of Aspartyl Proteinases Revealed by a Pepstatin Fragment Binding to Penicillopepsin Authors: James, M.N.G. / Sielecki, A. / Salituro, F. / Rich, D.H. / Hofmann, T. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ppk.cif.gz 1ppk.cif.gz | 81.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ppk.ent.gz pdb1ppk.ent.gz | 59.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ppk.json.gz 1ppk.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pp/1ppk https://data.pdbj.org/pub/pdb/validation_reports/pp/1ppk ftp://data.pdbj.org/pub/pdb/validation_reports/pp/1ppk ftp://data.pdbj.org/pub/pdb/validation_reports/pp/1ppk | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: CIS PROLINE - PRO E 134 / 2: THE REGION FROM SER E 277 - SER E 281 IS POORLY ORDERED. / 3: CIS PROLINE - PRO E 315 | ||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 1 types, 1 molecules E
| #1: Protein | Mass: 33468.809 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Penicillium janthinellum (fungus) / References: UniProt: P00798, penicillopepsin Penicillium janthinellum (fungus) / References: UniProt: P00798, penicillopepsin |
|---|
-Sugars , 2 types, 2 molecules 
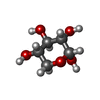

| #3: Sugar | ChemComp-MAN / |
|---|---|
| #4: Sugar | ChemComp-HSY / |
-Non-polymers , 4 types, 278 molecules 

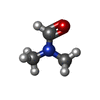


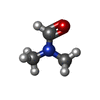

| #2: Chemical | ChemComp-IVV / |
|---|---|
| #5: Chemical | ChemComp-SO4 / |
| #6: Chemical | ChemComp-DMF / |
| #7: Water | ChemComp-HOH / |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.99 Å3/Da / Density % sol: 38.2 % | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Temperature: 18-22 ℃ / pH: 4.4 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | Highest resolution: 1.8 Å |
| Reflection | *PLUS Highest resolution: 1.8 Å / Lowest resolution: 45 Å / Num. obs: 24917 / Num. measured all: 29429 / Rmerge(I) obs: 0.033 |
- Processing
Processing
| Software | Name: PROLSQ / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.8→8 Å / σ(F): 3 /
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: PROLSQ / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.132 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller
























 PDBj
PDBj








