[English] 日本語
 Yorodumi
Yorodumi- PDB-1pfd: THE SOLUTION STRUCTURE OF HIGH PLANT PARSLEY [2FE-2S] FERREDOXIN,... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1pfd | ||||||
|---|---|---|---|---|---|---|---|
| Title | THE SOLUTION STRUCTURE OF HIGH PLANT PARSLEY [2FE-2S] FERREDOXIN, NMR, 18 STRUCTURES | ||||||
 Components Components | FERREDOXIN | ||||||
 Keywords Keywords | ELECTRON TRANSPORT / [2FE-2S] FERREDOXIN / SOLUTION STRUCTURE / PARAMAGNETISM / NUCLEAR RELAXATION | ||||||
| Function / homology |  Function and homology information Function and homology informationferredoxin metabolic process / chloroplast stroma / electron transport chain / 2 iron, 2 sulfur cluster binding / electron transfer activity / metal ion binding Similarity search - Function | ||||||
| Biological species |  Petroselinum crispum (parsley) Petroselinum crispum (parsley) | ||||||
| Method | SOLUTION NMR / distance geometry | ||||||
 Authors Authors | Im, S.-C. / Liu, G. / Luchinat, C. / Sykes, A.G. / Bertini, I. | ||||||
 Citation Citation |  Journal: Eur.J.Biochem. / Year: 1998 Journal: Eur.J.Biochem. / Year: 1998Title: The solution structure of parsley [2Fe-2S]ferredoxin. Authors: Im, S.C. / Liu, G. / Luchinat, C. / Sykes, A.G. / Bertini, I. #1:  Journal: Biochemistry / Year: 1996 Journal: Biochemistry / Year: 1996Title: Structure of Synechococcus Elongatus [Fe2S2] Ferredoxin in Solution Authors: Baumann, B. / Sticht, H. / Scharpf, M. / Sutter, M. / Haehnel, W. / Rosch, P. #2:  Journal: Biochemistry / Year: 1995 Journal: Biochemistry / Year: 1995Title: 1H and 15N NMR Sequential Assignment, Secondary Structure, and Tertiary Fold of [2Fe-2S] Ferredoxin from Synechocystis Sp. Pcc 6803 Authors: Lelong, C. / Setif, P. / Bottin, H. / Andre, F. / Neumann, J.M. #3:  Journal: Biochemistry / Year: 1994 Journal: Biochemistry / Year: 1994Title: An NMR-Derived Model for the Solution Structure of Oxidized Putidaredoxin, a 2-Fe, 2-S Ferredoxin from Pseudomonas Authors: Pochapsky, T.C. / Ye, X.M. / Ratnaswamy, G. / Lyons, T.A. #4:  Journal: Biochemistry / Year: 1990 Journal: Biochemistry / Year: 1990Title: Multinuclear Magnetic Resonance Studies of the 2Fe(Dot)2Sferredoxin from Anabaena Species Strain Pcc 7120. 1. Sequence-Specific Hydrogen-1 Resonance Assignments and Secondary Structure in ...Title: Multinuclear Magnetic Resonance Studies of the 2Fe(Dot)2Sferredoxin from Anabaena Species Strain Pcc 7120. 1. Sequence-Specific Hydrogen-1 Resonance Assignments and Secondary Structure in Solution of the Oxidized Form Authors: Oh, B.H. / Markley, J.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1pfd.cif.gz 1pfd.cif.gz | 493.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1pfd.ent.gz pdb1pfd.ent.gz | 409.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1pfd.json.gz 1pfd.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1pfd_validation.pdf.gz 1pfd_validation.pdf.gz | 376.8 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1pfd_full_validation.pdf.gz 1pfd_full_validation.pdf.gz | 485.9 KB | Display | |
| Data in XML |  1pfd_validation.xml.gz 1pfd_validation.xml.gz | 27.1 KB | Display | |
| Data in CIF |  1pfd_validation.cif.gz 1pfd_validation.cif.gz | 46.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pf/1pfd https://data.pdbj.org/pub/pdb/validation_reports/pf/1pfd ftp://data.pdbj.org/pub/pdb/validation_reports/pf/1pfd ftp://data.pdbj.org/pub/pdb/validation_reports/pf/1pfd | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 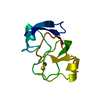
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 10496.383 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Petroselinum crispum (parsley) / Organ: LEAVES / References: UniProt: Q7M1S1, EC: 1.7.7.2 Petroselinum crispum (parsley) / Organ: LEAVES / References: UniProt: Q7M1S1, EC: 1.7.7.2 |
|---|---|
| #2: Chemical | ChemComp-FES / |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: MODEL NUMBER 1 WAS DETERMINED USING 2D NMR SPECTROSCOPY |
- Sample preparation
Sample preparation
| Details | Contents: WATER |
|---|---|
| Sample conditions | Ionic strength: 0.10 M / pH: 7 / Pressure: NORMAL / Temperature: 298 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| Software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software |
| ||||||||||||||||||||
| Refinement | Method: distance geometry / Software ordinal: 1 | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: LEAST TARGET FUNCTION / Conformers calculated total number: 50 / Conformers submitted total number: 18 |
 Movie
Movie Controller
Controller









 PDBj
PDBj










 AMBER
AMBER