[English] 日本語
 Yorodumi
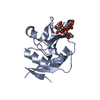
Yorodumi- PDB-1npq: structure of a rhodamine-labeled N-domain Troponin C mutant (Ca2+... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1npq | ||||||
|---|---|---|---|---|---|---|---|
| Title | structure of a rhodamine-labeled N-domain Troponin C mutant (Ca2+ saturated) in complex with skeletal Troponin I 115-131 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | STRUCTURAL PROTEIN / Troponin C- Troponin I complex / bifunctional rhodamine labeled Toponin C | ||||||
| Function / homology |  Function and homology information Function and homology informationtroponin T binding / Striated Muscle Contraction / troponin complex / skeletal muscle contraction / cardiac muscle contraction / actin binding / calcium ion binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method | SOLUTION NMR / simulated annealing using torsion angle dynamics, cartesian dynamics with the program CNS 1.1 | ||||||
| Model type details | minimized average | ||||||
 Authors Authors | Mercier, P. / Ferguson, R.E. / Irving, M. / Corrie, J.E.T. / Trentham, D.R. / Sykes, B.D. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2003 Journal: Biochemistry / Year: 2003Title: NMR Structure of a Bifunctional Rhodamine Labeled N-Domain of Troponin C Complexed with the Regulatory "Switch" Peptide from Troponin I: Implications for in Situ Fluorescence Studies in Muscle Fibers Authors: Mercier, P. / Ferguson, R.E. / Irving, M. / Corrie, J.E.T. / Trentham, D.R. / Sykes, B.D. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1npq.cif.gz 1npq.cif.gz | 664.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1npq.ent.gz pdb1npq.ent.gz | 555.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1npq.json.gz 1npq.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/np/1npq https://data.pdbj.org/pub/pdb/validation_reports/np/1npq ftp://data.pdbj.org/pub/pdb/validation_reports/np/1npq ftp://data.pdbj.org/pub/pdb/validation_reports/np/1npq | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 9932.144 Da / Num. of mol.: 1 / Fragment: TnC, residues 1-90 / Mutation: E56C, E63C Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #2: Protein/peptide | Mass: 1890.303 Da / Num. of mol.: 1 / Fragment: switch peptide, residues 115-131 / Source method: obtained synthetically Details: This sequence occurs naturally in Oryctolagus cuniculus (rabit) References: UniProt: P02643 |
| #3: Chemical |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||
| NMR details | Text: The structure was determined using triple-resonance spectroscopy. The chemical shifts for TnI115-131 were obtained from a 2D_15N/13C_filtered-DIPSI experiment. Intramolecular NOEs for TnI115- ...Text: The structure was determined using triple-resonance spectroscopy. The chemical shifts for TnI115-131 were obtained from a 2D_15N/13C_filtered-DIPSI experiment. Intramolecular NOEs for TnI115-131 were obtained from a 2D_15N/13C_filtered-NOESY experiment. |
- Sample preparation
Sample preparation
| Details | Contents: 320 mM KCl, 10 mM imidazole, 1.3% NaN3, pH 6.5, ~1mM sNTnC.2Ca2+.TnI115-131.BR56-63 Solvent system: 90% H2O/10% D2O |
|---|---|
| Sample conditions | Ionic strength: 320 mM KCl / pH: 6.5 / Pressure: ambiant / Temperature: 303 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing using torsion angle dynamics, cartesian dynamics with the program CNS 1.1 Software ordinal: 1 Details: calcium restraints were introduced only during the 2nd cooling stage using cartesian dynamics. See table 3 of the reference paper for a detailed description of the distance and dihedral restraints. | ||||||||||||||||||||||||
| NMR representative | Selection criteria: minimized average structure | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 100 / Conformers submitted total number: 21 |
 Movie
Movie Controller
Controller










 PDBj
PDBj



 NMRPipe
NMRPipe