[English] 日本語
 Yorodumi
Yorodumi- PDB-1njn: The crystal structure of the 50S Large ribosomal subunit from Dei... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1njn | ||||||
|---|---|---|---|---|---|---|---|
| Title | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with the antibiotic sparsomycin | ||||||
 Components Components | 23S ribosomal RNA | ||||||
 Keywords Keywords | RIBOSOME / Ribosomes / tRNA / puromycin / sparsomycin / peptidyl-transferase / peptide bond formation | ||||||
| Function / homology | SPARSOMYCIN / : / RNA / RNA (> 10) / RNA (> 100) / RNA (> 1000) Function and homology information Function and homology information | ||||||
| Biological species |  Deinococcus radiodurans (radioresistant) Deinococcus radiodurans (radioresistant) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / DIFFERENCE FOURIER MAPS / Resolution: 3.7 Å SYNCHROTRON / DIFFERENCE FOURIER MAPS / Resolution: 3.7 Å | ||||||
 Authors Authors | Bashan, A. / Agmon, I. / Zarivatch, R. / Schluenzen, F. / Harms, J.M. / Berisio, R. / Bartels, H. / Hansen, H.A. / Yonath, A. | ||||||
 Citation Citation |  Journal: Mol.Cell / Year: 2003 Journal: Mol.Cell / Year: 2003Title: Structural basis of the ribosomal machinery for Peptide bond formation, translocation, and nascent chain progression Authors: Bashan, A. / Agmon, I. / Zarivatch, R. / Schluenzen, F. / Harms, J.M. / Berisio, R. / Bartels, H. / Franceschi, F. / Auerbach, T. / Hansen, H.A. / Kossoy, E. / Kessler, M. / Yonath, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1njn.cif.gz 1njn.cif.gz | 1.3 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1njn.ent.gz pdb1njn.ent.gz | 1002 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1njn.json.gz 1njn.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/nj/1njn https://data.pdbj.org/pub/pdb/validation_reports/nj/1njn ftp://data.pdbj.org/pub/pdb/validation_reports/nj/1njn ftp://data.pdbj.org/pub/pdb/validation_reports/nj/1njn | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1njmC  1njoC  1njpC  1nkwS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
| #1: RNA chain | Mass: 933405.000 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Deinococcus radiodurans (radioresistant) / References: GenBank: 6460405 Deinococcus radiodurans (radioresistant) / References: GenBank: 6460405 |
|---|---|
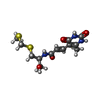
| #2: Chemical | ChemComp-SPS / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4 Å3/Da / Density % sol: 64 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 7.8 Details: ETHANOL, DIMETHYLHEXANEDIOL, MGCL2, KCL, HEPES, NH4CL, SPARSOMYCIN, pH 7.80 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 18 ℃ / pH: 7.8 / Method: vapor diffusionDetails: Harms, J.M., (2001) Cell (Cambridge,Mass.), 107, 679. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 90 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-4 / Wavelength: 0.9331 / Beamline: ID14-4 / Wavelength: 0.9331 |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Sep 1, 2001 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9331 Å / Relative weight: 1 |
| Reflection | Resolution: 3.7→40 Å / Num. obs: 206787 / % possible obs: 81.1 % / Observed criterion σ(I): -3 / Rsym value: 0.126 / Net I/σ(I): 5.1 |
| Reflection shell | Resolution: 3.7→3.76 Å / Mean I/σ(I) obs: 1.7 / Rsym value: 0.37 / % possible all: 80 |
| Reflection | *PLUS Lowest resolution: 40 Å / Rmerge(I) obs: 0.126 |
| Reflection shell | *PLUS % possible obs: 80 % / Rmerge(I) obs: 0.37 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: DIFFERENCE FOURIER MAPS Starting model: PDB ENTRY 1NKW Resolution: 3.7→15 Å / Rfactor Rfree error: 0.003 / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: ENGH & HUBER
| ||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.7→15 Å
| ||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor Rfree: 0.3117 / Rfactor Rwork: 0.271 | ||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS Type: c_angle_d |
 Movie
Movie Controller
Controller

















 PDBj
PDBj