+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1na1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of HRV14 when complexed with Pleconaril | |||||||||
 Components Components | (Coat protein ...) x 4 | |||||||||
 Keywords Keywords | VIRUS / HRV14 / human rhinovirus 14 / Pleconaril / Icosahedral virus | |||||||||
| Function / homology |  Function and homology information Function and homology informationlysis of host organelle involved in viral entry into host cell / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity ...lysis of host organelle involved in viral entry into host cell / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  Human rhinovirus 14 Human rhinovirus 14 | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.3 Å MOLECULAR REPLACEMENT / Resolution: 3.3 Å | |||||||||
 Authors Authors | Zhang, Y. / Simpson, A.A. / Bator, C.M. / Chakravarty, S. / Pevear, D.C. / Skochko, G.A. / Tull, T.M. / Diana, G. / Rossmann, M.G. | |||||||||
 Citation Citation |  Journal: J.Virol. / Year: 2004 Journal: J.Virol. / Year: 2004Title: Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds Authors: Zhang, Y. / Simpson, A.A. / Ledford, R.M. / Bator, C.M. / Chakravarty, S. / Skochko, G.A. / Demenczuk, T.M. / Watanyar, A. / Pevear, D.C. / Rossmann, M.G. | |||||||||
| History |
| |||||||||
| Remark 999 | AUTHORS STATE THAT RESIDUE 170 (CHAIN B) CAN BE ILE OR LEU, AS SEEN IN OTHER DATABASE REFERENCE SEQUENCES. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1na1.cif.gz 1na1.cif.gz | 176.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1na1.ent.gz pdb1na1.ent.gz | 138.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1na1.json.gz 1na1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/na/1na1 https://data.pdbj.org/pub/pdb/validation_reports/na/1na1 ftp://data.pdbj.org/pub/pdb/validation_reports/na/1na1 ftp://data.pdbj.org/pub/pdb/validation_reports/na/1na1 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | x 60
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | x 5
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | x 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | x 20
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: (Hermann–Mauguin notation: 532 / Schoenflies symbol: I (icosahedral)) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
-Coat protein ... , 4 types, 4 molecules ABCD
| #1: Protein | Mass: 32560.549 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Human rhinovirus 14 / Genus: Rhinovirus / Cell line: HELA CELLS / Species: Human rhinovirus B / References: UniProt: P03303 Human rhinovirus 14 / Genus: Rhinovirus / Cell line: HELA CELLS / Species: Human rhinovirus B / References: UniProt: P03303 |
|---|---|
| #2: Protein | Mass: 28501.361 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Human rhinovirus 14 / Genus: Rhinovirus / Cell line: HELA CELLS / Species: Human rhinovirus B / References: UniProt: P03303 Human rhinovirus 14 / Genus: Rhinovirus / Cell line: HELA CELLS / Species: Human rhinovirus B / References: UniProt: P03303 |
| #3: Protein | Mass: 26236.754 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Human rhinovirus 14 / Genus: Rhinovirus / Cell line: HELA CELLS / Species: Human rhinovirus B / References: UniProt: P03303 Human rhinovirus 14 / Genus: Rhinovirus / Cell line: HELA CELLS / Species: Human rhinovirus B / References: UniProt: P03303 |
| #4: Protein | Mass: 7183.863 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Human rhinovirus 14 / Genus: Rhinovirus / Cell line: HELA CELLS / Species: Human rhinovirus B / References: UniProt: P03303 Human rhinovirus 14 / Genus: Rhinovirus / Cell line: HELA CELLS / Species: Human rhinovirus B / References: UniProt: P03303 |
-Non-polymers , 2 types, 243 molecules 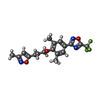


| #5: Chemical | ChemComp-W11 / |
|---|---|
| #6: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 7.5 Details: HEPES, NaCl, CaCl2, PEG8K, pH 7.5, VAPOR DIFFUSION, HANGING DROP, temperature 293K | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 14-BM-C / Wavelength: 1 Å / Beamline: 14-BM-C / Wavelength: 1 Å |
| Detector | Type: FUJI / Detector: IMAGE PLATE / Date: Sep 14, 2001 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 3.3→20 Å / Num. all: 320952 / Num. obs: 320952 / % possible obs: 77.8 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Rsym value: 0.164 |
| Reflection shell | Highest resolution: 3.3 Å / % possible all: 77.8 |
| Reflection | *PLUS Num. measured all: 545390 / Rmerge(I) obs: 0.164 |
| Reflection shell | *PLUS % possible obs: 68.2 % / Rmerge(I) obs: 0.477 |
- Processing
Processing
| Software |
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 3.3→20 Å / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber MOLECULAR REPLACEMENT / Resolution: 3.3→20 Å / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| |||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.3→20 Å
| |||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller

















 PDBj
PDBj


