[English] 日本語
 Yorodumi
Yorodumi- PDB-1mzt: NMR structure of the fd bacteriophage pVIII coat protein in lipid... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1mzt | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR structure of the fd bacteriophage pVIII coat protein in lipid bilayer membranes | ||||||
 Components Components | major coat protein pVIII | ||||||
 Keywords Keywords | VIRAL PROTEIN / fd coat protein / membrane-bound / pVIII | ||||||
| Function / homology | Phage major coat protein, Gp8 / Bacteriophage M13, G8P, capsid domain superfamily / Capsid protein G8P / helical viral capsid / host cell membrane / membrane / Capsid protein G8P Function and homology information Function and homology information | ||||||
| Biological species |  Enterobacteria phage fd (virus) Enterobacteria phage fd (virus) | ||||||
| Method | SOLID-STATE NMR / Calculation of bond orientations, dihedral angles from solid-state NMR restraints, back-calculation of NMR data from oriented structure | ||||||
 Authors Authors | Marassi, F.M. / Opella, S.J. | ||||||
 Citation Citation |  Journal: Protein Sci. / Year: 2003 Journal: Protein Sci. / Year: 2003Title: Simultaneous assignment and structure determination of a membrane protein from NMR orientational restraints Authors: Marassi, F.M. / Opella, S.J. #1:  Journal: J.Biomol.NMR / Year: 2002 Journal: J.Biomol.NMR / Year: 2002Title: Using Pisa pies to resolve ambiguities in angular constraints from PISEMA spectra of aligned proteins Authors: Marassi, F.M. / Opella, S.J. | ||||||
| History |
| ||||||
| Remark 999 | SEQUENCE AUTHOR PROVIDED COORDINATES FOR THE PROTEIN BACKBONE ONLY. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1mzt.cif.gz 1mzt.cif.gz | 14.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1mzt.ent.gz pdb1mzt.ent.gz | 6.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1mzt.json.gz 1mzt.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1mzt_validation.pdf.gz 1mzt_validation.pdf.gz | 244.6 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1mzt_full_validation.pdf.gz 1mzt_full_validation.pdf.gz | 244.4 KB | Display | |
| Data in XML |  1mzt_validation.xml.gz 1mzt_validation.xml.gz | 2.1 KB | Display | |
| Data in CIF |  1mzt_validation.cif.gz 1mzt_validation.cif.gz | 2.2 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mz/1mzt https://data.pdbj.org/pub/pdb/validation_reports/mz/1mzt ftp://data.pdbj.org/pub/pdb/validation_reports/mz/1mzt ftp://data.pdbj.org/pub/pdb/validation_reports/mz/1mzt | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 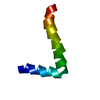
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 5244.000 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Enterobacteria phage fd (virus) / Genus: Inovirus / Species: Enterobacteria phage M13 / References: UniProt: P69539 Enterobacteria phage fd (virus) / Genus: Inovirus / Species: Enterobacteria phage M13 / References: UniProt: P69539 |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLID-STATE NMR |
|---|---|
| NMR experiment | Type: 1H/15N PISEMA |
| NMR details | Text: The structure was determined from one uniformly and four selectively 15N-labeled samples. |
- Sample preparation
Sample preparation
| Details | Contents: 8 mg 15N-labeled fd coat protein, 64 mg POPC, 16 mg POPG, lipid bilayers oriented on glass slides Solvent system: Lipid bilayers hydrated in water |
|---|---|
| Sample conditions | Ionic strength: 0 / pH: 7 / Pressure: ambient / Temperature: 296 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Chemagnetics CMX / Manufacturer: Chemagnetics / Model: CMX / Field strength: 400 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: Calculation of bond orientations, dihedral angles from solid-state NMR restraints, back-calculation of NMR data from oriented structure Software ordinal: 1 Details: Structure calculated from 40 chemical shift and 40 dipolar coupling restraints | ||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 6 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller







 PDBj
PDBj