+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1m4w | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Thermophilic b-1,4-xylanase from Nonomuraea flexuosa | |||||||||
 Components Components | endoxylanase | |||||||||
 Keywords Keywords | HYDROLASE / xylanase / glycosyl hydrolase / family 11 / thermostability | |||||||||
| Function / homology |  Function and homology information Function and homology informationendo-1,4-beta-xylanase activity / endo-1,4-beta-xylanase / xylan catabolic process / polysaccharide binding Similarity search - Function | |||||||||
| Biological species |  Thermopolyspora flexuosa (bacteria) Thermopolyspora flexuosa (bacteria) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å MOLECULAR REPLACEMENT / Resolution: 2.1 Å | |||||||||
 Authors Authors | Hakulinen, N. / Turunen, O. / Janis, J. / Leisola, M. / Rouvinen, J. | |||||||||
 Citation Citation |  Journal: Eur.J.Biochem. / Year: 2003 Journal: Eur.J.Biochem. / Year: 2003Title: Three-dimensional structures of thermophilic beta-1,4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa. Comparison of twelve xylanases in relation to their thermal stability. Authors: Hakulinen, N. / Turunen, O. / Leisola, M. / Rouvinen, J. | |||||||||
| History |
| |||||||||
| Remark 999 | SEQUENCE At the time of processing, there was no database sequence available for this protein. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1m4w.cif.gz 1m4w.cif.gz | 58.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1m4w.ent.gz pdb1m4w.ent.gz | 40.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1m4w.json.gz 1m4w.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/m4/1m4w https://data.pdbj.org/pub/pdb/validation_reports/m4/1m4w ftp://data.pdbj.org/pub/pdb/validation_reports/m4/1m4w ftp://data.pdbj.org/pub/pdb/validation_reports/m4/1m4w | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1h1aC  1enxS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 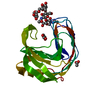
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 21802.447 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Thermopolyspora flexuosa (bacteria) / Production host: Thermopolyspora flexuosa (bacteria) / Production host:  Hypocrea jecorina (fungus) Hypocrea jecorina (fungus)References: GenBank: 23337128, UniProt: Q8GMV7*PLUS, endo-1,4-beta-xylanase |
|---|---|
| #2: Polysaccharide | alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1- ...alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source |
| #3: Chemical | ChemComp-ACT / |
| #4: Chemical | ChemComp-GOL / |
| #5: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 6 Details: 0.8 M Ammonium sulphate, 0.1 M Sodium Acetate, pH 6.0, VAPOR DIFFUSION, HANGING DROP, temperature 293K | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 120 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å |
| Detector | Type: RIGAKU / Detector: IMAGE PLATE / Date: Oct 20, 2000 / Details: MSC Confocal Blue |
| Radiation | Monochromator: graphite / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.1→99 Å / Num. obs: 8541 / % possible obs: 97.6 % / Redundancy: 5.7 % / Biso Wilson estimate: 23 Å2 / Rsym value: 0.107 / Net I/σ(I): 17.9 |
| Reflection shell | Resolution: 2.1→2.18 Å / Rsym value: 0.293 / % possible all: 92.8 |
| Reflection | *PLUS Num. measured all: 48587 / Rmerge(I) obs: 0.107 |
| Reflection shell | *PLUS % possible obs: 92.8 % / Rmerge(I) obs: 0.293 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1ENX Resolution: 2.1→99 Å / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.1→99 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller











 PDBj
PDBj






