[English] 日本語
 Yorodumi
Yorodumi- PDB-1lkk: HUMAN P56-LCK TYROSINE KINASE SH2 DOMAIN IN COMPLEX WITH THE PHOS... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1lkk | ||||||
|---|---|---|---|---|---|---|---|
| Title | HUMAN P56-LCK TYROSINE KINASE SH2 DOMAIN IN COMPLEX WITH THE PHOSPHOTYROSYL PEPTIDE AC-PTYR-GLU-GLU-ILE (PYEEI PEPTIDE) | ||||||
 Components Components |
| ||||||
 Keywords Keywords | COMPLEX (TYROSINE KINASE/PEPTIDE) / COMPLEX (TYROSINE KINASE-PEPTIDE) / COMPLEX (TYROSINE KINASE-PEPTIDE) complex | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of lymphocyte activation / positive regulation of leukocyte cell-cell adhesion / CD27 signaling pathway / regulation of regulatory T cell differentiation / gamma-delta T cell differentiation / positive regulation of gamma-delta T cell differentiation / Fc-gamma receptor signaling pathway / FLT3 signaling through SRC family kinases / protein antigen binding / Nef Mediated CD4 Down-regulation ...regulation of lymphocyte activation / positive regulation of leukocyte cell-cell adhesion / CD27 signaling pathway / regulation of regulatory T cell differentiation / gamma-delta T cell differentiation / positive regulation of gamma-delta T cell differentiation / Fc-gamma receptor signaling pathway / FLT3 signaling through SRC family kinases / protein antigen binding / Nef Mediated CD4 Down-regulation / intracellular zinc ion homeostasis / CD4 receptor binding / positive regulation of heterotypic cell-cell adhesion / Nef and signal transduction / Co-stimulation by CD28 / Interleukin-2 signaling / CD28 dependent Vav1 pathway / peptidyl-tyrosine autophosphorylation / Regulation of KIT signaling / leukocyte migration / phospholipase activator activity / Co-inhibition by CTLA4 / CD8 receptor binding / Translocation of ZAP-70 to Immunological synapse / Phosphorylation of CD3 and TCR zeta chains / pericentriolar material / protein serine/threonine phosphatase activity / PECAM1 interactions / positive regulation of T cell receptor signaling pathway / hemopoiesis / Generation of second messenger molecules / RHOH GTPase cycle / immunological synapse / T cell differentiation / Co-inhibition by PD-1 / CD28 dependent PI3K/Akt signaling / T cell receptor binding / phosphatidylinositol 3-kinase binding / phospholipase binding / GPVI-mediated activation cascade / T cell costimulation / positive regulation of intrinsic apoptotic signaling pathway / phosphotyrosine residue binding / release of sequestered calcium ion into cytosol / cell surface receptor protein tyrosine kinase signaling pathway / SH2 domain binding / peptidyl-tyrosine phosphorylation / Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants / T cell activation / B cell receptor signaling pathway / non-membrane spanning protein tyrosine kinase activity / non-specific protein-tyrosine kinase / Signaling by SCF-KIT / platelet activation / positive regulation of T cell activation / Constitutive Signaling by Aberrant PI3K in Cancer / cell-cell junction / DAP12 signaling / Downstream TCR signaling / PIP3 activates AKT signaling / T cell receptor signaling pathway / ATPase binding / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / protein tyrosine kinase activity / protein phosphatase binding / protein phosphorylation / intracellular signal transduction / membrane raft / response to xenobiotic stimulus / signaling receptor binding / positive regulation of gene expression / protein kinase binding / extracellular exosome / ATP binding / identical protein binding / plasma membrane / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1 Å X-RAY DIFFRACTION / Resolution: 1 Å | ||||||
 Authors Authors | Tong, L. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1996 Journal: J.Mol.Biol. / Year: 1996Title: Crystal structures of the human p56lck SH2 domain in complex with two short phosphotyrosyl peptides at 1.0 A and 1.8 A resolution. Authors: Tong, L. / Warren, T.C. / King, J. / Betageri, R. / Rose, J. / Jakes, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1lkk.cif.gz 1lkk.cif.gz | 94.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1lkk.ent.gz pdb1lkk.ent.gz | 71.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1lkk.json.gz 1lkk.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lk/1lkk https://data.pdbj.org/pub/pdb/validation_reports/lk/1lkk ftp://data.pdbj.org/pub/pdb/validation_reports/lk/1lkk ftp://data.pdbj.org/pub/pdb/validation_reports/lk/1lkk | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 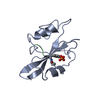
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 11985.371 Da / Num. of mol.: 1 / Fragment: SH2 DOMAIN Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Production host: Homo sapiens (human) / Production host:  |
|---|---|
| #2: Protein/peptide | Mass: 658.591 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source |
| #3: Chemical | ChemComp-ACE / |
| #4: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.96 Å3/Da / Density % sol: 37.28 % | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS pH: 7 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | *PLUS Highest resolution: 1 Å / Num. obs: 48786 / % possible obs: 91 % / Num. measured all: 170160 / Rmerge(I) obs: 0.07 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1→30 Å / σ(I): -3 Details: THE REFINEMENT WAS CARRIED OUT AGAINST F**2. REFLECTIONS THAT HAVE NEGATIVE INTENSITIES ARE INCLUDED IN THE REFINEMENT. RESIDUES 170 - 176 HAVE WEAK ELECTRON DENSITY. RESIDUES LEU 122, HIS ...Details: THE REFINEMENT WAS CARRIED OUT AGAINST F**2. REFLECTIONS THAT HAVE NEGATIVE INTENSITIES ARE INCLUDED IN THE REFINEMENT. RESIDUES 170 - 176 HAVE WEAK ELECTRON DENSITY. RESIDUES LEU 122, HIS 148, THR 177, ARG 184, AND ARG 196 HAVE WEAK SIDE CHAIN ELECTRON DENSITY. ANISOTROPIC TEMPERATURE FACTORS WERE REFINED FOR ALL NON-HYDROGEN ATOMS. HYDROGEN ATOMS FOR THE SH2 DOMAIN AND THE INHIBITOR WERE INCLUDED IN THE REFINEMENT, WITH RIDING ISOTROPIC TEMPERATURE FACTOR VALUES. HYDROGEN ATOMS ON THE WATER MOLECULES WERE NOT INCLUDED.
| |||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1→30 Å
| |||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS Type: s_angle_deg / Dev ideal: 1.7 |
 Movie
Movie Controller
Controller









 PDBj
PDBj


















