[English] 日本語
 Yorodumi
Yorodumi- PDB-1kyu: AP-2 CLATHRIN ADAPTOR ALPHA-APPENDAGE IN COMPLEX WITH EPS15 DPF P... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1kyu | ||||||
|---|---|---|---|---|---|---|---|
| Title | AP-2 CLATHRIN ADAPTOR ALPHA-APPENDAGE IN COMPLEX WITH EPS15 DPF PEPTIDE | ||||||
 Components Components |
| ||||||
 Keywords Keywords | ENDOCYTOSIS/EXOCYTOSIS / PROTEIN-PEPTIDE COMPLEX / ENDOCYTOSIS / ENDOCYTOSIS-EXOCYTOSIS COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationNegative regulation of MET activity / EGFR downregulation / LDL clearance / WNT5A-dependent internalization of FZD2, FZD5 and ROR2 / WNT5A-dependent internalization of FZD4 / Trafficking of GluR2-containing AMPA receptors / Retrograde neurotrophin signalling / ubiquitin-dependent endocytosis / VLDLR internalisation and degradation / Recycling pathway of L1 ...Negative regulation of MET activity / EGFR downregulation / LDL clearance / WNT5A-dependent internalization of FZD2, FZD5 and ROR2 / WNT5A-dependent internalization of FZD4 / Trafficking of GluR2-containing AMPA receptors / Retrograde neurotrophin signalling / ubiquitin-dependent endocytosis / VLDLR internalisation and degradation / Recycling pathway of L1 / Golgi to endosome transport / clathrin coat of coated pit / postsynaptic endocytic zone / AP-2 adaptor complex / postsynaptic neurotransmitter receptor internalization / Cargo recognition for clathrin-mediated endocytosis / membrane coat / Clathrin-mediated endocytosis / clathrin coat assembly / clathrin-cargo adaptor activity / clathrin-dependent endocytosis / MHC class II antigen presentation / endocytic recycling / clathrin-coated vesicle / regulation of hematopoietic stem cell differentiation / aggresome / ciliary membrane / positive regulation of receptor recycling / polyubiquitin modification-dependent protein binding / synaptic vesicle endocytosis / vesicle-mediated transport / receptor-mediated endocytosis of virus by host cell / clathrin-coated pit / Neutrophil degranulation / basal plasma membrane / secretory granule / ubiquitin binding / intracellular protein transport / SH3 domain binding / cytoplasmic side of plasma membrane / kinase binding / disordered domain specific binding / regulation of cell population proliferation / presynapse / regulation of protein localization / cytoplasmic vesicle / early endosome membrane / postsynapse / apical plasma membrane / protein domain specific binding / calcium ion binding / lipid binding / protein kinase binding / glutamatergic synapse / identical protein binding / plasma membrane / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.8 Å MOLECULAR REPLACEMENT / Resolution: 1.8 Å | ||||||
 Authors Authors | Brett, T.J. / Traub, L.M. / Fremont, D.H. | ||||||
 Citation Citation |  Journal: Structure / Year: 2002 Journal: Structure / Year: 2002Title: Accessory protein recruitment motifs in clathrin-mediated endocytosis. Authors: Brett, T.J. / Traub, L.M. / Fremont, D.H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1kyu.cif.gz 1kyu.cif.gz | 66.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1kyu.ent.gz pdb1kyu.ent.gz | 48.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1kyu.json.gz 1kyu.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ky/1kyu https://data.pdbj.org/pub/pdb/validation_reports/ky/1kyu ftp://data.pdbj.org/pub/pdb/validation_reports/ky/1kyu ftp://data.pdbj.org/pub/pdb/validation_reports/ky/1kyu | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1ky6C  1ky7C  1kydC  1kyfC  1qtpS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 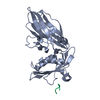
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 27828.676 Da / Num. of mol.: 1 / Fragment: C-TERMINAL APPENDAGE (EAR) RESIDUES 701-938 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #2: Protein/peptide | Mass: 650.701 Da / Num. of mol.: 1 / Fragment: RESIDUES 628-632 / Source method: obtained synthetically Details: The peptide was chemically synthesized. The sequence is a naturally occuring sequence in EPS15 (MOUSE) References: UniProt: P42567 |
| #3: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.02 Å3/Da / Density % sol: 39.21 % | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS pH: 6.8 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 110 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU300 / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU RU300 / Wavelength: 1.5418 Å |
| Detector | Type: RIGAKU RAXIS IV / Detector: IMAGE PLATE / Date: Jun 28, 2000 / Details: YALE MIRRORS |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.8→50 Å / Num. all: 20976 / Num. obs: 20976 / % possible obs: 93.1 % / Observed criterion σ(F): 0 / Observed criterion σ(I): -3 / Biso Wilson estimate: 18.5 Å2 / Rsym value: 0.052 / Net I/σ(I): 18.3 |
| Reflection shell | Resolution: 1.8→1.86 Å / Mean I/σ(I) obs: 2.8 / Rsym value: 0.382 / % possible all: 89.3 |
| Reflection | *PLUS Num. measured all: 86516 / Rmerge(I) obs: 0.052 |
| Reflection shell | *PLUS % possible obs: 89.3 % / Rmerge(I) obs: 0.382 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1QTP Resolution: 1.8→19.96 Å / Rfactor Rfree error: 0.013 / Data cutoff high absF: 1127447.1 / Data cutoff high rms absF: 1127447.1 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 56.4235 Å2 / ksol: 0.390539 e/Å3 | ||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 28.3 Å2
| ||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→19.96 Å
| ||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.8→1.91 Å / Rfactor Rfree error: 0.042 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS % reflection Rfree: 5 % / Rfactor all: 0.188 / Rfactor obs: 0.187 / Rfactor Rfree: 0.223 / Rfactor Rwork: 0.187 | ||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rfree: 0.377 / Rfactor Rwork: 0.293 / Rfactor obs: 0.293 |
 Movie
Movie Controller
Controller










 PDBj
PDBj













