+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1koe | ||||||
|---|---|---|---|---|---|---|---|
| Title | ENDOSTATIN | ||||||
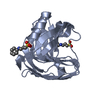
 Components Components | ENDOSTATIN | ||||||
 Keywords Keywords | ANGIOGENESIS INHIBITOR / COLLAGEN XVIII / NON-COLLAGENEOUS DOMAIN / HEPARIN-BINDING DOMAIN | ||||||
| Function / homology |  Function and homology information Function and homology informationActivation of Matrix Metalloproteinases / Collagen biosynthesis and modifying enzymes / Collagen chain trimerization / Collagen degradation / Assembly of collagen fibrils and other multimeric structures / vascular endothelial cell proliferation / blood vessel endothelial cell migration / Integrin cell surface interactions / endothelial cell morphogenesis / collagen trimer ...Activation of Matrix Metalloproteinases / Collagen biosynthesis and modifying enzymes / Collagen chain trimerization / Collagen degradation / Assembly of collagen fibrils and other multimeric structures / vascular endothelial cell proliferation / blood vessel endothelial cell migration / Integrin cell surface interactions / endothelial cell morphogenesis / collagen trimer / endothelial cell apoptotic process / positive regulation of vascular endothelial cell proliferation / basement membrane / positive regulation of endothelial cell apoptotic process / positive regulation of blood vessel endothelial cell migration / extracellular matrix organization / : / angiogenesis / cell adhesion / extracellular space / metal ion binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MIRAS, SOLVENT FLATTENING AT 2.2 A RESOLUTION / Resolution: 1.5 Å MIRAS, SOLVENT FLATTENING AT 2.2 A RESOLUTION / Resolution: 1.5 Å | ||||||
 Authors Authors | Hohenester, E. / Sasaki, T. / Olsen, B.R. / Timpl, R. | ||||||
 Citation Citation |  Journal: EMBO J. / Year: 1998 Journal: EMBO J. / Year: 1998Title: Crystal structure of the angiogenesis inhibitor endostatin at 1.5 A resolution. Authors: Hohenester, E. / Sasaki, T. / Olsen, B.R. / Timpl, R. #1:  Journal: Cell(Cambridge,Mass.) / Year: 1997 Journal: Cell(Cambridge,Mass.) / Year: 1997Title: Endostatin: An Endogenous Inhibitor of Angiogenesis and Tumor Growth Authors: O'Reilly, M.S. / Boehm, T. / Shing, Y. / Fukai, N. / Vasios, G. / Lane, W.S. / Flynn, E. / Birkhead, J.R. / Olsen, B.R. / Folkman, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1koe.cif.gz 1koe.cif.gz | 47.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1koe.ent.gz pdb1koe.ent.gz | 32.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1koe.json.gz 1koe.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1koe_validation.pdf.gz 1koe_validation.pdf.gz | 418.6 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1koe_full_validation.pdf.gz 1koe_full_validation.pdf.gz | 418.9 KB | Display | |
| Data in XML |  1koe_validation.xml.gz 1koe_validation.xml.gz | 10.2 KB | Display | |
| Data in CIF |  1koe_validation.cif.gz 1koe_validation.cif.gz | 13.1 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ko/1koe https://data.pdbj.org/pub/pdb/validation_reports/ko/1koe ftp://data.pdbj.org/pub/pdb/validation_reports/ko/1koe ftp://data.pdbj.org/pub/pdb/validation_reports/ko/1koe | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 18950.355 Da / Num. of mol.: 1 Fragment: C-TERMINAL 184 RESIDUES OF ALPHA1 CHAIN OF COLLAGEN XVIII Source method: isolated from a genetically manipulated source Details: N-TERMINAL ALA-PRO-LEU-ALA SEQUENCE INTRODUCED BY THE CLONING PROCESS Source: (gene. exp.)   Homo sapiens (human) / References: UniProt: P39061 Homo sapiens (human) / References: UniProt: P39061 |
|---|---|
| #2: Water | ChemComp-HOH / |
| Has protein modification | Y |
| Sequence details | NUMBERING SCHEME IS BASED ON THE POSITION IN THE CARBOXY-TERMINAL NON-COLLAGENEOUS (NC1) DOMAIN OF ...NUMBERING SCHEME IS BASED ON THE POSITION IN THE CARBOXY-TERMINAL NON-COLLAGENEO |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.95 Å3/Da / Density % sol: 37 % Description: THREE DERIVATIVES WERE USED, Crystal grow | pH: 5.3 / Details: pH 5.3 | Crystal grow | *PLUS Method: vapor diffusion, hanging dropDetails: drop solution was mixed with an equal volume of reservoir solution Components of the solutions | *PLUS
|
|---|
-Data collection
| Diffraction | Mean temperature: 293 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SRS SRS  / Beamline: PX9.6 / Wavelength: 0.87 / Beamline: PX9.6 / Wavelength: 0.87 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Nov 17, 1997 |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.87 Å / Relative weight: 1 |
| Reflection | Resolution: 1.5→25 Å / Num. obs: 26471 / % possible obs: 99.3 % / Observed criterion σ(I): 0 / Redundancy: 5.9 % / Rmerge(I) obs: 0.075 / Net I/σ(I): 5.5 |
| Reflection shell | Resolution: 1.5→1.55 Å / Redundancy: 5.9 % / Rmerge(I) obs: 0.18 / Mean I/σ(I) obs: 3.9 / % possible all: 99.2 |
| Reflection | *PLUS Num. measured all: 159011 |
| Reflection shell | *PLUS % possible obs: 99.2 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MIRAS, SOLVENT FLATTENING AT 2.2 A RESOLUTION MIRAS, SOLVENT FLATTENING AT 2.2 A RESOLUTIONResolution: 1.5→8 Å / Data cutoff high absF: 1000000 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED INDIVIDUAL B-F / Cross valid method: FREE R-FACTOR / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 16 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati sigma a obs: 0.12 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.5→8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.5→1.55 Å / Total num. of bins used: 10
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.1 / Classification: refinement X-PLOR / Version: 3.1 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor obs: 0.249 |
 Movie
Movie Controller
Controller












 PDBj
PDBj








