| 登録情報 | データベース: PDB / ID: 1kj0
|
|---|
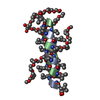
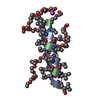
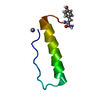
| タイトル | SOLUTION STRUCTURE OF THE SMALL SERINE PROTEASE INHIBITOR SGTI |
|---|
 要素 要素 | SERINE PROTEASE INHIBITOR I |
|---|
 キーワード キーワード | HYDROLASE / serine protease inhibition / inhibitor specificity |
|---|
| 機能・相同性 | Protease inhibitor with pacifastin repeats / Pacifastin domain / Pacifastin domain superfamily / Pacifastin inhibitor (LCMII) / Pacifastin domain profile. / serine-type endopeptidase inhibitor activity / extracellular region / Serine protease inhibitor I/II 機能・相同性情報 機能・相同性情報 |
|---|
| 手法 | 溶液NMR / simulated annealing |
|---|
 データ登録者 データ登録者 | Gaspari, Z. / Patthy, A. / Graf, L. / Perczel, A. |
|---|
 引用 引用 |  ジャーナル: Eur.J.Biochem. / 年: 2002 ジャーナル: Eur.J.Biochem. / 年: 2002
タイトル: Comparative structure analysis of proteinase inhibitors from the desert locust, Schistocerca gregaria.
著者: Gaspari, Z. / Patthy, A. / Graf, L. / Perczel, A. |
|---|
| 履歴 | | 登録 | 2001年12月4日 | 登録サイト: RCSB / 処理サイト: RCSB |
|---|
| 改定 1.0 | 2001年12月12日 | Provider: repository / タイプ: Initial release |
|---|
| 改定 1.1 | 2008年4月27日 | Group: Version format compliance |
|---|
| 改定 1.2 | 2011年7月13日 | Group: Version format compliance |
|---|
| 改定 1.3 | 2022年2月23日 | Group: Database references / Derived calculations
カテゴリ: database_2 / pdbx_struct_assembly / pdbx_struct_oper_list
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
| 改定 1.4 | 2024年10月30日 | Group: Data collection / Structure summary
カテゴリ: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / pdbx_entry_details / pdbx_modification_feature |
|---|
|
|---|
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 データ登録者
データ登録者 引用
引用 ジャーナル: Eur.J.Biochem. / 年: 2002
ジャーナル: Eur.J.Biochem. / 年: 2002 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 1kj0.cif.gz
1kj0.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb1kj0.ent.gz
pdb1kj0.ent.gz PDB形式
PDB形式 1kj0.json.gz
1kj0.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード https://data.pdbj.org/pub/pdb/validation_reports/kj/1kj0
https://data.pdbj.org/pub/pdb/validation_reports/kj/1kj0 ftp://data.pdbj.org/pub/pdb/validation_reports/kj/1kj0
ftp://data.pdbj.org/pub/pdb/validation_reports/kj/1kj0 リンク
リンク 集合体
集合体
 要素
要素 試料調製
試料調製 解析
解析 ムービー
ムービー コントローラー
コントローラー











 PDBj
PDBj X-PLOR
X-PLOR