[English] 日本語
 Yorodumi
Yorodumi- PDB-1kci: Crystal Structure of 9-amino-N-[2-(4-morpholinyl)ethyl]-4-acridin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1kci | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of 9-amino-N-[2-(4-morpholinyl)ethyl]-4-acridinecarboxamide Bound to d(CGTACG)2 | ||||||||||||||||||
 Components Components | 5'-D(* Keywords KeywordsDNA / intercalator / aminoacridinecarboxamide | Function / homology | Chem-DRC / DNA |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / ISOMORPHOUS / Resolution: 1.8 Å X-RAY DIFFRACTION / ISOMORPHOUS / Resolution: 1.8 Å  Authors AuthorsAdams, A. / Guss, J.M. / Denny, W.A. / Wakelin, L.P.G. |  Citation Citation Journal: Nucleic Acids Res. / Year: 2002 Journal: Nucleic Acids Res. / Year: 2002Title: Crystal structure of 9-amino-N-[2-(4-morpholinyl)ethyl]-4-acridinecarboxamide bound to d(CGTACG)2: implications for structure-activity relationships of acridinecarboxamide topoisomerase poisons. Authors: Adams, A. / Guss, J.M. / Denny, W.A. / Wakelin, L.P. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1kci.cif.gz 1kci.cif.gz | 15.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1kci.ent.gz pdb1kci.ent.gz | 8.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1kci.json.gz 1kci.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/kc/1kci https://data.pdbj.org/pub/pdb/validation_reports/kc/1kci ftp://data.pdbj.org/pub/pdb/validation_reports/kc/1kci ftp://data.pdbj.org/pub/pdb/validation_reports/kc/1kci | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 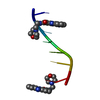
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: DNA chain | Mass: 1809.217 Da / Num. of mol.: 1 / Source method: obtained synthetically | ||
|---|---|---|---|
| #2: Chemical | | #3: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.75 Å3/Da / Density % sol: 55.25 % | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 285 K / Method: vapor diffusion, sitting drop / pH: 6 Details: cacodylate, spermine, KCl, MgCl2, MPD, pH 6.0, VAPOR DIFFUSION, SITTING DROP, temperature 285K | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 110 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å |
| Detector | Type: RIGAKU RAXIS II / Detector: IMAGE PLATE / Date: Aug 1, 2000 / Details: focusing mirror |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.8→40 Å / Num. all: 1951 / Num. obs: 1951 / % possible obs: 99.8 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 7.7 % / Rsym value: 0.094 / Net I/σ(I): 21.5 |
| Reflection shell | Resolution: 1.8→1.86 Å / Redundancy: 6.6 % / Mean I/σ(I) obs: 5 / Num. unique all: 195 / Rsym value: 0.35 / % possible all: 100 |
| Reflection | *PLUS Lowest resolution: 40 Å / Num. obs: 1955 / Num. measured all: 30670 / Rmerge(I) obs: 0.094 |
| Reflection shell | *PLUS % possible obs: 100 % / Rmerge(I) obs: 0.351 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: ISOMORPHOUS Starting model: NDB entry DD0015 Resolution: 1.8→40 Å / Isotropic thermal model: isotropic / Cross valid method: Rfree / σ(F): 0 / σ(I): 0 / Stereochemistry target values: SHELX 2000 DNA dictioary
| ||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 28 Å2 | ||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→40 Å
| ||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: SHELXL-97 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 1.8 Å / Lowest resolution: 40 Å / σ(F): 0 | ||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller



 PDBj
PDBj