[English] 日本語
 Yorodumi
Yorodumi- PDB-1kb0: Crystal Structure of Quinohemoprotein Alcohol Dehydrogenase from ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1kb0 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of Quinohemoprotein Alcohol Dehydrogenase from Comamonas testosteroni | ||||||
 Components Components | quinohemoprotein alcohol dehydrogenase | ||||||
 Keywords Keywords | OXIDOREDUCTASE / beta-propeller fold / cytochrome c | ||||||
| Function / homology |  Function and homology information Function and homology informationalcohol dehydrogenase (azurin) / oxidoreductase activity, acting on CH-OH group of donors / outer membrane-bounded periplasmic space / electron transfer activity / heme binding / calcium ion binding / membrane Similarity search - Function | ||||||
| Biological species |  Comamonas testosteroni (bacteria) Comamonas testosteroni (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.44 Å MOLECULAR REPLACEMENT / Resolution: 1.44 Å | ||||||
 Authors Authors | Rozeboom, H.J. / Oubrie, A. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2002 Journal: J.Biol.Chem. / Year: 2002Title: Crystal structure of quinohemoprotein alcohol dehydrogenase from Comamonas testosteroni: structural basis for substrate oxidation and electron transfer. Authors: Oubrie, A. / Rozeboom, H.J. / Kalk, K.H. / Huizinga, E.G. / Dijkstra, B.W. #1:  Journal: ACTA CRYSTALLOGR.,SECT.D / Year: 2001 Journal: ACTA CRYSTALLOGR.,SECT.D / Year: 2001Title: Crystallization of quinohaemoprotein alcohol dehydrogenase from Comamonas testosteroni: crystals with unique optical properties. Authors: Oubrie, A. / Huizinga, E.G. / Rozeboom, H.J. / Kalk, K.H. / de Jong, G.A. / Duine, J.A. / Dijkstra, B.W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1kb0.cif.gz 1kb0.cif.gz | 173.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1kb0.ent.gz pdb1kb0.ent.gz | 130.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1kb0.json.gz 1kb0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/kb/1kb0 https://data.pdbj.org/pub/pdb/validation_reports/kb/1kb0 ftp://data.pdbj.org/pub/pdb/validation_reports/kb/1kb0 ftp://data.pdbj.org/pub/pdb/validation_reports/kb/1kb0 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1eee  1flgS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 73438.461 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Comamonas testosteroni (bacteria) Comamonas testosteroni (bacteria)References: UniProt: Q46444, Oxidoreductases; Acting on the CH-OH group of donors; With unknown physiological acceptors |
|---|
-Non-polymers , 6 types, 1023 molecules 

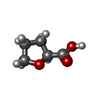
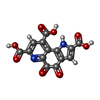







| #2: Chemical | ChemComp-CA / | ||
|---|---|---|---|
| #3: Chemical | ChemComp-HEC / | ||
| #4: Chemical | ChemComp-TFB / | ||
| #5: Chemical | ChemComp-PQQ / | ||
| #6: Chemical | | #7: Water | ChemComp-HOH / | |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.2 Å3/Da / Density % sol: 44 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 5.7 Details: PEG 6000, MES, pH 5.7, VAPOR DIFFUSION, HANGING DROP, temperature 277K | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Details: Oubrie, A., (2001) ACTA CRYSTALLOGR.,SECT.D, 57, 1732.PH range low: 5.7 / PH range high: 5.5 | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: X11 / Wavelength: 0.9076 Å / Beamline: X11 / Wavelength: 0.9076 Å | |||||||||
| Detector | Type: MAR scanner 345 mm plate / Detector: IMAGE PLATE / Date: Sep 27, 1998 | |||||||||
| Radiation | Protocol: OSCILLATION / Scattering type: x-ray | |||||||||
| Radiation wavelength | Wavelength: 0.9076 Å / Relative weight: 1 | |||||||||
| Reflection | Resolution: 1.44→50 Å / Num. obs: 111086 / % possible obs: 96.2 % / Redundancy: 2.2 % / Biso Wilson estimate: 19 Å2 / Rmerge(I) obs: 0.057 / Rsym value: 0.057 / Net I/σ(I): 16 | |||||||||
| Reflection shell | Resolution: 1.44→1.46 Å / Redundancy: 1.7 % / Rmerge(I) obs: 0.34 / Mean I/σ(I) obs: 2 / Rsym value: 0.34 / % possible all: 87.7 | |||||||||
| Reflection shell | *PLUS % possible obs: 87.7 % / Rmerge(I) obs: 0.34 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1EEE (now known as 1FLG) Resolution: 1.44→29.63 Å / Rfactor Rfree error: 0.002 / Data cutoff high absF: 1721072.94 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber / Details: HOH 492 belongs to conformation A of ASP 657.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 64.9357 Å2 / ksol: 0.377884 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 21.5 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.44→29.63 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.44→1.53 Å / Rfactor Rfree error: 0.008 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: CNS / Version: 1 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Lowest resolution: 50 Å / σ(F): 0 / % reflection Rfree: 5 % / Rfactor obs: 0.16 / Rfactor Rwork: 0.16 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 21.5 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rfree: 0.259 / % reflection Rfree: 5.1 % / Rfactor Rwork: 0.228 |
 Movie
Movie Controller
Controller












 PDBj
PDBj













