[English] 日本語
 Yorodumi
Yorodumi- PDB-1k3g: NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1k3g | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | ||||||
 Components Components | cytochrome c-553 | ||||||
 Keywords Keywords | ELECTRON TRANSPORT / C-553 / HEME / CYTOCHROME / BACILLUS PASTEURII / ELECTRON TRANSFER | ||||||
| Function / homology |  Function and homology information Function and homology informationelectron transfer activity / iron ion binding / heme binding / plasma membrane Similarity search - Function | ||||||
| Biological species |  Sporosarcina pasteurii (bacteria) Sporosarcina pasteurii (bacteria) | ||||||
| Method | SOLUTION NMR / torsion angle dynamics, restrained energy minimization | ||||||
 Authors Authors | Banci, L. / Bertini, I. / Ciurli, S. / Dikiy, A. / Dittmer, J. / Rosato, A. / Sciara, G. / Thompsett, A.R. | ||||||
 Citation Citation |  Journal: Chembiochem / Year: 2002 Journal: Chembiochem / Year: 2002Title: NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria. Authors: Banci, L. / Bertini, I. / Ciurli, S. / Dikiy, A. / Dittmer, J. / Rosato, A. / Sciara, G. / Thompsett, A.R. #1:  Journal: Biochemistry / Year: 2000 Journal: Biochemistry / Year: 2000Title: Crystal Structure of Oxidized Bacillus pasteurii cytochrome c-553 at 0.97-A Resolution Authors: Benini, S. / Gonzalez, A. / Rypniewski, W.R. / Wilson, K.S. / Van Beeumen, J.J. / Ciurli, S. #2:  Journal: J.Biol.Inorg.Chem. / Year: 1998 Journal: J.Biol.Inorg.Chem. / Year: 1998Title: Modulation of Bacillus pasteurii Cytochrome c553 Reduction Potential by Structural and Solution Parameters Authors: Benini, S. / Borsari, M. / Ciurli, S. / Dikiy, A. / Lamborghini, M. | ||||||
| History |
|
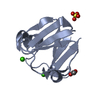
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1k3g.cif.gz 1k3g.cif.gz | 676.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1k3g.ent.gz pdb1k3g.ent.gz | 576.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1k3g.json.gz 1k3g.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/k3/1k3g https://data.pdbj.org/pub/pdb/validation_reports/k3/1k3g ftp://data.pdbj.org/pub/pdb/validation_reports/k3/1k3g ftp://data.pdbj.org/pub/pdb/validation_reports/k3/1k3g | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 7115.937 Da / Num. of mol.: 1 / Fragment: residues 22-92 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Sporosarcina pasteurii (bacteria) / Plasmid: pEC86 / Production host: Sporosarcina pasteurii (bacteria) / Plasmid: pEC86 / Production host:  |
|---|---|
| #2: Chemical | ChemComp-HEC / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 1-3 mM oxidized cytochrome c-553 in 10 mM phosphate buffer Solvent system: 90%H2O+10%D2O; 100% D2O |
|---|---|
| Sample conditions | Ionic strength: 10 mM phosphate buffer / pH: 7.5 / Pressure: Ambient / Temperature: 288 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiation wavelength | Relative weight: 1 | ||||||||||||||||||||
| NMR spectrometer |
|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: torsion angle dynamics, restrained energy minimization Software ordinal: 1 Details: PSEUDOCONTACT SHIFTS WERE INCLUDED AS CONSTRAINTS BY MEANS OF MODIFIED DYANA AND SANDER MODULES (PSEUDODYANA, PSEUDOREM) (BANCI ET AL., 1997) | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: target function / Conformers calculated total number: 400 / Conformers submitted total number: 30 |
 Movie
Movie Controller
Controller











 PDBj
PDBj









 Amber
Amber