[English] 日本語
 Yorodumi
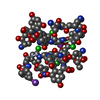
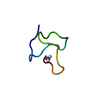
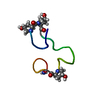
Yorodumi- PDB-1jlo: Solution Structure of the Noncompetitive Skeletal Muscle Nicotini... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1jlo | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution Structure of the Noncompetitive Skeletal Muscle Nicotinic Acetylcholine Receptor Antagonist Psi-conotoxin PIIIE | ||||||
 Components Components | PSI-CONOTOXIN PIIIE | ||||||
 Keywords Keywords | TOXIN / MULTIPLE DISULFIDE BONDS / AMIDATED C-TERMINUS | ||||||
| Function / homology | Conotoxin / Conotoxin / host cell postsynaptic membrane / acetylcholine receptor inhibitor activity / ion channel inhibitor activity / toxin activity / extracellular region / Psi-conotoxin PIIIE Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / distance geometry molecular dynamics relaxation matrix distance geometry simulated annealing | ||||||
 Authors Authors | Van Wagoner, R.M. / Ireland, C.M. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2003 Journal: Biochemistry / Year: 2003Title: An Improved Solution Structure for psi-Conotoxin Piiie Authors: Van Wagoner, R.M. / Ireland, C.M. #1:  Journal: Biochemistry / Year: 1998 Journal: Biochemistry / Year: 1998Title: Three-dimensional Solution Structure of Conotoxin psi-PIIIE, an Acetylcholine Gated Ion Channel Antagonist Authors: Mitchell, S.S. / Shon, K.J. / Foster, M.P. / Davis, D.R. / Olivera, B.M. / Ireland, C.M. #2:  Journal: Biochemistry / Year: 1997 Journal: Biochemistry / Year: 1997Title: A Noncompetitive Peptide Inhibitor of the Nicotinic Acetylcholine Receptor from Conus purpurascens Venom Authors: Shon, K.J. / Grilley, M. / Jacobsen, R. / Cartier, G.E. / Hopkins, C. / Gray, W.R. / Watkins, M. / Hillyard, D.R. / Rivier, J. / Torres, J. / Yoshikami, D. / Olivera, B.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1jlo.cif.gz 1jlo.cif.gz | 86.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1jlo.ent.gz pdb1jlo.ent.gz | 59.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1jlo.json.gz 1jlo.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1jlo_validation.pdf.gz 1jlo_validation.pdf.gz | 353.8 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1jlo_full_validation.pdf.gz 1jlo_full_validation.pdf.gz | 392.8 KB | Display | |
| Data in XML |  1jlo_validation.xml.gz 1jlo_validation.xml.gz | 7.9 KB | Display | |
| Data in CIF |  1jlo_validation.cif.gz 1jlo_validation.cif.gz | 12 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/jl/1jlo https://data.pdbj.org/pub/pdb/validation_reports/jl/1jlo ftp://data.pdbj.org/pub/pdb/validation_reports/jl/1jlo ftp://data.pdbj.org/pub/pdb/validation_reports/jl/1jlo | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 2727.181 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: Solid phase chemical synthesis was used to incorporate 3-13C-labeled cysteine at positions 4 and 21. The sequence is the same as the native peptide from CONUS PURPURASCENS (PURPLE CONE). References: UniProt: P56529 |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: This structure was determined using isotopic filtration and isotope-selected 1H observation in addition to standard homonuclear techniques. |
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
| |||||||||||||||
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiation wavelength | Relative weight: 1 | |||||||||||||||
| NMR spectrometer |
|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: distance geometry molecular dynamics relaxation matrix distance geometry simulated annealing Software ordinal: 1 Details: These structures are based on 566 NOE-derived distance restraints, 9 restraints on phi derived from J-coupling, and 13 restraints on chi1 determined from J-coupling and NOE volumes. | ||||||||||||||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: back calculated data agree with experimental NOESY spectrum, structures with the least restraint violations, structures with the lowest energy Conformers calculated total number: 50 / Conformers submitted total number: 13 |
 Movie
Movie Controller
Controller








 PDBj
PDBj