+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1jes | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of a Copper-Mediated Base Pair in DNA | ||||||||||||||||||
 Components Components | 5'-D(* Keywords KeywordsDNA / Cu / copper / Z-DNA / dodecamer / duplex / unnatural / designed | Function / homology | COPPER (II) ION / DNA / DNA (> 10) |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 1.5 Å MAD / Resolution: 1.5 Å  Authors AuthorsAtwell, S. / Meggers, E. / Spraggon, G. / Schultz, P.G. |  Citation Citation Journal: J.Am.Chem.Soc. / Year: 2001 Journal: J.Am.Chem.Soc. / Year: 2001Title: Structure of a Copper-Mediated Base Pair in DNA Authors: Atwell, S. / Meggers, E. / Spraggon, G. / Schultz, P.G. #1:  Journal: J.Am.Chem.Soc. / Year: 2000 Journal: J.Am.Chem.Soc. / Year: 2000Title: A Novel Copper-Mediated DNA Base Pair Authors: Meggers, E. / Holland, P.L. / Tolman, W.B. / Romesberg, F.E. / Schultz, P.G. #2:  Journal: Science / Year: 1981 Journal: Science / Year: 1981Title: Left-handed double helical DNA: variations in the backbone conformation Authors: Wang, A.G.-J. / Quigley, G.J. / Kolpak, F.J. / van Der Marel, G. / van Boom, J.H. / Rich, A. #3:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1981 Journal: Proc.Natl.Acad.Sci.USA / Year: 1981Title: Structure of a B-DNA dodecamer: conformation and dynamics Authors: Drew, H.R. / Wing, R.M. / Takano, T. / Broka, C. / Tanaka, S. / Itakura, K. / Dickerson, R.E. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1jes.cif.gz 1jes.cif.gz | 25.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1jes.ent.gz pdb1jes.ent.gz | 17.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1jes.json.gz 1jes.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/je/1jes https://data.pdbj.org/pub/pdb/validation_reports/je/1jes ftp://data.pdbj.org/pub/pdb/validation_reports/je/1jes ftp://data.pdbj.org/pub/pdb/validation_reports/je/1jes | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 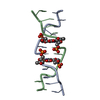
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 3648.371 Da / Num. of mol.: 2 / Source method: obtained synthetically #2: Chemical | #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.82 Å3/Da / Density % sol: 32.43 % | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 7 Details: 10% MPD, 40 mM Na Cacodylate, 12 mM Spermine tetra-HCl, 80 mM NaCl, 20 mM MgCl2, pH 7.0, VAPOR DIFFUSION, HANGING DROP, temperature 298K | ||||||||||||||||||||||||
| Components of the solutions |
|
-Data collection
| Diffraction | Mean temperature: 140 K | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 5.0.2 / Wavelength: 1.37991,1.37791,1.0 / Beamline: 5.0.2 / Wavelength: 1.37991,1.37791,1.0 | ||||||||||||
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Apr 6, 2001 / Details: Curved Si (111) | ||||||||||||
| Radiation | Protocol: MAD / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||
| Radiation wavelength |
| ||||||||||||
| Reflection | Resolution: 1.5→50 Å / Num. all: 47272 / Num. obs: 47190 / % possible obs: 98.2 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 5.5 % / Biso Wilson estimate: 14.058 Å2 / Rsym value: 0.096 / Net I/σ(I): 46.7 | ||||||||||||
| Reflection shell | Resolution: 1.49→1.54 Å / Rmerge(I) obs: 0.686 / % possible all: 84.1 | ||||||||||||
| Reflection | *PLUS Num. obs: 8531 / Num. measured all: 47272 / Rmerge(I) obs: 0.096 | ||||||||||||
| Reflection shell | *PLUS % possible obs: 84.1 % / Rmerge(I) obs: 0.686 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MAD MADStarting model: initial map fit with mononucleotides Resolution: 1.5→50 Å / Cross valid method: free r / σ(F): 0 / σ(I): 0 Stereochemistry target values: G. Parkinson, J. Vojtechovsky, L. Clowney, A.T. Brunger, H.M. Berman, New Parameters for the Refinement of Nucleic Acid Containing Structures, Acta Cryst. D, 52, 57-64 (1996). Details: The fofc maps indicated an alternate conformation for the phosphate of B23, and so the structure was refined with two conformations for the phosphate of B23 (excepting the O5') and most of ...Details: The fofc maps indicated an alternate conformation for the phosphate of B23, and so the structure was refined with two conformations for the phosphate of B23 (excepting the O5') and most of residue B22 (excepting O5'), each with 0.5 occupancy.
| |||||||||||||||||||||||||
| Displacement parameters |
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.5→50 Å
| |||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||
| Software | *PLUS Name: CNS / Classification: refinement | |||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 1.5 Å / Lowest resolution: 50 Å / Num. reflection obs: 8531 / σ(F): 0 / % reflection Rfree: 8 % / Rfactor obs: 0.169 | |||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller













 PDBj
PDBj










































