[English] 日本語
 Yorodumi
Yorodumi- PDB-1ia2: Candida albicans dihydrofolate reductase complexed with dihydro-n... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ia2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Candida albicans dihydrofolate reductase complexed with dihydro-nicotinamide-adenine-dinucleotide phosphate (NADPH) and 5-[(4-METHYLPHENYL)SULFANYL]-2,4-QUINAZOLINEDIAMINE (GW578) | ||||||
 Components Components | DIHYDROFOLATE REDUCTASE | ||||||
 Keywords Keywords | OXIDOREDUCTASE / ANTIFUNGAL TARGET / REDUCTASE | ||||||
| Function / homology |  Function and homology information Function and homology informationdihydrofolate metabolic process / dihydrofolate reductase / dihydrofolate reductase activity / folic acid metabolic process / tetrahydrofolate biosynthetic process / one-carbon metabolic process / NADP binding / mitochondrion Similarity search - Function | ||||||
| Biological species |  Candida albicans (yeast) Candida albicans (yeast) | ||||||
| Method |  X-RAY DIFFRACTION / DIRECT REPLACEMENT / Resolution: 1.82 Å X-RAY DIFFRACTION / DIRECT REPLACEMENT / Resolution: 1.82 Å | ||||||
 Authors Authors | Whitlow, M. / Howard, A.J. / Kuyper, L.F. | ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2001 Journal: J.Med.Chem. / Year: 2001Title: X-ray Crystal Structures of Candida albicans Dihydrofolate Reductase: High Resolution Ternary Complexes in Which the Dihydronicotinamide Moiety of NADPH is Displaced by an inhibitor Authors: Whitlow, M. / Howard, A.J. / Stewart, D. / Hardman, K.D. / Chan, J.H. / Baccanari, D.P. / Tansik, R.L. / Hong, J.S. / Kuyper, L.F. #1:  Journal: J.Biol.Chem. / Year: 1997 Journal: J.Biol.Chem. / Year: 1997Title: X-ray Crystallographic Studies of Candida albicans Dihydrofolate Reductase. High resolution structures of the holoenzyme and an inhibited ternary complex. Authors: Whitlow, M. / Howard, A.J. / Stewart, D. / Hardman, K.D. / Kuyper, L.F. / Baccanari, D.P. / Fling, M.E. / Tansik, R.L. #2:  Journal: J.Med.Chem. / Year: 1995 Journal: J.Med.Chem. / Year: 1995Title: Selective Inhibitors of Candida albicans Dihydrofolate Reductase: Activity and Selectivity of 5-(arylthio)-2,4-diaminoquinazolines Authors: Chan, J.H. / Hong, J.S. / Kuyper, L.F. / Baccanari, D.P. / Joyner, S.S. / Tansik, R.L. / Boytos, C.M. / Rudolph, S.K. #3:  Journal: J.Biol.Chem. / Year: 1989 Journal: J.Biol.Chem. / Year: 1989Title: Characterization of Candida Albicans Dihydrofolate Reductase Authors: Baccanari, D.P. / Tansik, R.L. / Joyner, S.S. / Fling, M.E. / Smith, P.L. / Freisheim, J.H. | ||||||
| History |
| ||||||
| Remark 99 | HET GROUPS NDP 193, TQ4 194, AND MES 201 ARE ASSOCIATED WITH PROTEIN CHAIN A. HET GROUPS NDP 195, ...HET GROUPS NDP 193, TQ4 194, AND MES 201 ARE ASSOCIATED WITH PROTEIN CHAIN A. HET GROUPS NDP 195, TQ4 196, AND MES 202 ARE ASSOCIATED WITH PROTEIN CHAIN B. | ||||||
| Remark 600 | HETEROGEN THE INHIBITION CONSTANT (IC50) FOR 5-[(4-METHYLPHENYL) SULFANYL]-2,4-QUINAZOLINEDIAMINE ... HETEROGEN THE INHIBITION CONSTANT (IC50) FOR 5-[(4-METHYLPHENYL) SULFANYL]-2,4-QUINAZOLINEDIAMINE (GW578) IS 23 NANOMOLAR USING THE FOLLOWING ASSAY (SEE REFERENCE 0 & 3). C. ALBICANS DHFR ASSAY WAS PERFORMED IN 0.1 M IMIDAZOLE CHLORIDE BUFFER, PH 6.4, WITH 12 MM MERCAPTOETHANOL, 60 MM NADPH AND 45 MM DIHYDROFOLIC ACID IN A FINAL VOLUME OF 1 ML AT 303K. IC50 IS THE CONCENTRATION OF INHIBITOR THAT DECREASES THE VELOCITY OF THE STANDARD ASSAY BY 50%. THE ENZYME (0.2 NM), NADPH, AND VARYING CONCENTRATIONS OF INHIBITOR WERE PREINCUBATED FOR 2 MIN AT 30-C, AND THE REACTION WAS INITIATED BY THE ADDITION OF DIHYDROFOLIC ACID. STEADY STATE VELOCITIES WERE MEASURED, AND IC50 VALUES WERE CALCULATED FROM A LINEAR REGRESSION PLOT OF THE PERCENTAGE INHIBITION VS THE LOGARITHM OF THE INHIBITOR CONCENTRATION. IC50 THE PRECISION OF THE DETERMINATION IS GENERALLY ABOUT A 30%. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ia2.cif.gz 1ia2.cif.gz | 107.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ia2.ent.gz pdb1ia2.ent.gz | 81.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ia2.json.gz 1ia2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ia/1ia2 https://data.pdbj.org/pub/pdb/validation_reports/ia/1ia2 ftp://data.pdbj.org/pub/pdb/validation_reports/ia/1ia2 ftp://data.pdbj.org/pub/pdb/validation_reports/ia/1ia2 | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 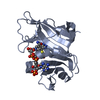 1ia1C  1ia3C  1ia4C  1ai9S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | Candida DHFR is active as a monomer. |
- Components
Components
| #1: Protein | Mass: 22194.527 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Candida albicans (yeast) / Gene: DFR1 / Plasmid: P1869 / Species (production host): Escherichia coli / Production host: Candida albicans (yeast) / Gene: DFR1 / Plasmid: P1869 / Species (production host): Escherichia coli / Production host:  #2: Chemical | #3: Chemical | #4: Chemical | #5: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.27 Å3/Da / Density % sol: 43 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: DIHYDRO-NICOTINAMIDE-ADENINE- DINUCLEOTIDE PHOSPHATE (NADPH), 5-[(4-METHYLPHENYL) SULFANYL]-2,4-QUINAZOLINEDIAMINE (GW578), PEG-3350, POTASSIUM 4-MORPHILINEETHANESULFONIC ACID (MES), ...Details: DIHYDRO-NICOTINAMIDE-ADENINE- DINUCLEOTIDE PHOSPHATE (NADPH), 5-[(4-METHYLPHENYL) SULFANYL]-2,4-QUINAZOLINEDIAMINE (GW578), PEG-3350, POTASSIUM 4-MORPHILINEETHANESULFONIC ACID (MES), DITHIOTHREITOL (DTT). A THREE-FOLD EXCESS OF GW578 AND THREE-FOLD EXCESS OF NADPH WAS ADDED TO THE C. ALBICANS DHFR SOLUTION AND LET STAND 277K OVERNIGHT. 17-20 MG/ML C. ALBICANS DHFR IN 50 UM GW578, 50 UM NADPH, 20 MM KMES, 1 MM DTT, PH 6.5 WAS MIXED WITH AN EQUAL PART OF 26 - 34% PEG-3350, THE RESERVOIR SOLUTION. , VAPOR DIFFUSION, HANGING DROP | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 4 ℃ | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 293 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: ELLIOTT GX-21 / Wavelength: 1.5418 ROTATING ANODE / Type: ELLIOTT GX-21 / Wavelength: 1.5418 |
| Detector | Type: XENTRONICS / Detector: AREA DETECTOR / Date: Feb 22, 1989 / Details: MONOCHROMATOR |
| Radiation | Monochromator: Huber graphite monochromator / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Highest resolution: 1.76 Å / Num. all: 137080 / Num. obs: 35749 / % possible obs: 93.5 % / Observed criterion σ(F): 0 / Observed criterion σ(I): -3 / Redundancy: 3.83 % / Biso Wilson estimate: 30.7 Å2 / Rmerge(I) obs: 0.0761 / Rsym value: 0.0761 / Net I/σ(I): 12.8 |
| Reflection shell | Resolution: 1.76→1.87 Å / Redundancy: 2.17 % / Rmerge(I) obs: 0.2983 / Mean I/σ(I) obs: 1.82 / Num. unique all: 4426 / Rsym value: 0.2983 / % possible all: 69.6 |
| Reflection shell | *PLUS % possible obs: 69.6 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: DIRECT REPLACEMENT Starting model: CANDIDA ALBICANS DHFR (PDB ENTRY 1AI9) Resolution: 1.82→10 Å Isotropic thermal model: Konnert, J.H. & Hendrickson, W.A. (1980) Acta Crystallogr. A A36, 344. σ(F): 2 / σ(I): 0 Stereochemistry target values: Hendrickson, W.A. (1985) Methods Enzymol. 115, 252-270. Details: Restrain least-squares procedure.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.82→10 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj