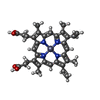
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1i55 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CYTOCHROME C (TUNA) WITH 2ZN:1FE MIXED-METAL PORPHYRINS | |||||||||
 Components Components | CYTOCHROME C | |||||||||
 Keywords Keywords | ELECTRON TRANSPORT / cytochrome c / electron transfer / zinc-porphyrin | |||||||||
| Function / homology |  Function and homology information Function and homology informationmitochondrial intermembrane space / electron transfer activity / heme binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Thunnus thynnus (Atlantic bluefin tuna) Thunnus thynnus (Atlantic bluefin tuna) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | |||||||||
 Authors Authors | Crane, B.R. / Tezcan, F.A. / Winkler, J.R. / Gray, H.B. | |||||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2001 Journal: Proc.Natl.Acad.Sci.USA / Year: 2001Title: Electron tunneling in protein crystals. Authors: Tezcan, F.A. / Crane, B.R. / Winkler, J.R. / Gray, H.B. | |||||||||
| History |
| |||||||||
| Remark 600 | HETEROGEN The residue 1104 of HEM and residue 1105 of ZNH are alternate conformers with occupancy ...HETEROGEN The residue 1104 of HEM and residue 1105 of ZNH are alternate conformers with occupancy of 0.39 and 0.61, respectively. In similar residue 1106 of HEM and residue 1107 of ZNH are alternate conformers with occupancy 0.39 and 0.61, respectively. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1i55.cif.gz 1i55.cif.gz | 67.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1i55.ent.gz pdb1i55.ent.gz | 48.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1i55.json.gz 1i55.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1i55_validation.pdf.gz 1i55_validation.pdf.gz | 705.6 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1i55_full_validation.pdf.gz 1i55_full_validation.pdf.gz | 710.7 KB | Display | |
| Data in XML |  1i55_validation.xml.gz 1i55_validation.xml.gz | 6.8 KB | Display | |
| Data in CIF |  1i55_validation.cif.gz 1i55_validation.cif.gz | 11.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/i5/1i55 https://data.pdbj.org/pub/pdb/validation_reports/i5/1i55 ftp://data.pdbj.org/pub/pdb/validation_reports/i5/1i55 ftp://data.pdbj.org/pub/pdb/validation_reports/i5/1i55 | HTTPS FTP |
-Related structure data
| Related structure data |  1i54C  3cytS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 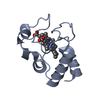
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 11390.076 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Thunnus thynnus (Atlantic bluefin tuna) / References: UniProt: P81459 Thunnus thynnus (Atlantic bluefin tuna) / References: UniProt: P81459#2: Chemical | #3: Chemical | #4: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.17 Å3/Da / Density % sol: 43.21 % | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / pH: 6 Details: (NH4)2SO4 NACL NaPi, pH 6.0, VAPOR DIFFUSION, SITTING DROP, temperature 298K | |||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop | |||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL9-2 / Wavelength: 1.282 Å / Beamline: BL9-2 / Wavelength: 1.282 Å |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Jul 22, 2000 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.282 Å / Relative weight: 1 |
| Reflection | Resolution: 2→3 Å / Num. all: 11058 / Num. obs: 19049 / % possible obs: 81.7 % / Observed criterion σ(F): 1 / Observed criterion σ(I): 1 / Redundancy: 1.7 % / Biso Wilson estimate: 17.2 Å2 / Rmerge(I) obs: 0.062 / Net I/σ(I): 17.1 |
| Reflection shell | Resolution: 2→2.07 Å / Redundancy: 1.5 % / Rmerge(I) obs: 0.13 / % possible all: 78.2 |
| Reflection | *PLUS Lowest resolution: 30 Å |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 3CYT Resolution: 2→19.64 Å / Rfactor Rfree error: 0.008 / Data cutoff high absF: 381490.79 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / σ(F): 0 / σ(I): 0 / Stereochemistry target values: Engh & Huber Details: Crystals contained a mixture of Zn- and Fe- porphyrins with occupancies determined from anomalous scattering measurements. Residues 1104 and 1105 and residues 1105 and 1106 were refined as ...Details: Crystals contained a mixture of Zn- and Fe- porphyrins with occupancies determined from anomalous scattering measurements. Residues 1104 and 1105 and residues 1105 and 1106 were refined as non-interacting porphyrins, reflecting the mixture of Zn and Fe cytc in the crystal lattice. No stereochemical restraints were placed on ZN or Fe interactions.
| ||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 27.98 Å2 / ksol: 0.398 e/Å3 | ||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 17.7 Å2
| ||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→19.64 Å
| ||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2→2.07 Å / Rfactor Rfree error: 0.027 / Total num. of bins used: 10
| ||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: CNS / Version: 1 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS σ(F): 0 / % reflection Rfree: 6.3 % | ||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 17.7 Å2 | ||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rfree: 0.278 / % reflection Rfree: 7.4 % / Rfactor Rwork: 0.26 |
 Movie
Movie Controller
Controller












 PDBj
PDBj