+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1hji | ||||||
|---|---|---|---|---|---|---|---|
| Title | BACTERIOPHAGE HK022 NUN-PROTEIN-NUTBOXB-RNA COMPLEX | ||||||
 Components Components |
| ||||||
 Keywords Keywords | BACTERIOPHAGE HK022 / TERMINATION / PEPTIDE-RNA-COMPLEX / PEPTIDE-RNA-RECOGNITION / PROTEIN/RNA | ||||||
| Function / homology | negative regulation of DNA-templated transcription, elongation / DNA-templated transcription termination / RNA / RNA (> 10) / Transcription termination factor nun Function and homology information Function and homology information | ||||||
| Biological species |  BACTERIOPHAGE LAMBDA (virus) BACTERIOPHAGE LAMBDA (virus) BACTERIOPHAGE HK022 (virus) BACTERIOPHAGE HK022 (virus) | ||||||
| Method | SOLUTION NMR / molecular dynamics | ||||||
 Authors Authors | Faber, C. / Schaerpf, M. / Becker, T. / Sticht, H. / Roesch, P. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2001 Journal: J.Biol.Chem. / Year: 2001Title: The Structure of the Coliphage Hk022 Nun Protein-Lambda-Phage Boxb RNA Complex. Implications for the Mechanism of Transcription Termination Authors: Faber, C. / Schaerpf, M. / Becker, T. / Sticht, H. / Roesch, P. #1:  Journal: Eur.J.Biochem. / Year: 2000 Journal: Eur.J.Biochem. / Year: 2000Title: Antitermination in Bacteriophage Lambda: The Structure of the N36 Peptide-Boxb RNA Complex Authors: Schaerpf, M. / Sticht, H. / Schweimer, K. / Boehm, M. / Hoffmann, S. / Roesch, P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1hji.cif.gz 1hji.cif.gz | 387.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1hji.ent.gz pdb1hji.ent.gz | 328.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1hji.json.gz 1hji.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hj/1hji https://data.pdbj.org/pub/pdb/validation_reports/hj/1hji ftp://data.pdbj.org/pub/pdb/validation_reports/hj/1hji ftp://data.pdbj.org/pub/pdb/validation_reports/hj/1hji | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 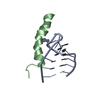
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 4853.994 Da / Num. of mol.: 1 / Fragment: BACTERIOPHAGE LAMBDA NUTBOXB-RNA Source method: isolated from a genetically manipulated source Details: NUTBOXB-RNA FROM THE NUTR-SEQUENCE / Source: (gene. exp.)  BACTERIOPHAGE LAMBDA (virus) / Production host: BACTERIOPHAGE LAMBDA (virus) / Production host:  |
|---|---|
| #2: Protein/peptide | Mass: 3167.657 Da / Num. of mol.: 1 / Fragment: N-TERMINAL BINDING-DOMAIN, RESIDUES 19-44 / Source method: obtained synthetically / Source: (synth.)  BACTERIOPHAGE HK022 (virus) / References: UniProt: P18683 BACTERIOPHAGE HK022 (virus) / References: UniProt: P18683 |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Sample conditions | Ionic strength: 90 mM / pH: 6.5 / Pressure: 1 atm / Temperature: 301.0 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker DRX / Manufacturer: Bruker / Model: DRX / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: molecular dynamics / Software ordinal: 1 Details: REFINEMENT DETAILS CAN BE FOUND IN THE JRNL CITATION ABOVE | ||||||||||||||||
| NMR ensemble | Conformer selection criteria: LOWEST ENERGY / Conformers calculated total number: 120 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller











 PDBj
PDBj





























 X-PLOR
X-PLOR