[English] 日本語
 Yorodumi
Yorodumi- PDB-2dq5: solution structure of the Mid1 B Box2 Chc(D/C)C2H2 Zinc-Binding D... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2dq5 | ||||||
|---|---|---|---|---|---|---|---|
| Title | solution structure of the Mid1 B Box2 Chc(D/C)C2H2 Zinc-Binding Domain: insights into an evolutionary conserved ring fold | ||||||
 Components Components | Midline-1 | ||||||
 Keywords Keywords | LIGASE / E3 Ligase / RING like / Zinc coordination | ||||||
| Function / homology |  Function and homology information Function and homology informationprotein localization to microtubule / positive regulation of stress-activated MAPK cascade / pattern specification process / negative regulation of microtubule depolymerization / microtubule associated complex / regulation of microtubule cytoskeleton organization / phosphoprotein binding / RING-type E3 ubiquitin transferase / microtubule cytoskeleton organization / centriolar satellite ...protein localization to microtubule / positive regulation of stress-activated MAPK cascade / pattern specification process / negative regulation of microtubule depolymerization / microtubule associated complex / regulation of microtubule cytoskeleton organization / phosphoprotein binding / RING-type E3 ubiquitin transferase / microtubule cytoskeleton organization / centriolar satellite / spindle / Interferon gamma signaling / transferase activity / microtubule binding / microtubule / ubiquitin protein ligase binding / enzyme binding / Golgi apparatus / protein homodimerization activity / zinc ion binding / identical protein binding / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | SOLUTION NMR / torsion angle dynamics, low target function | ||||||
 Authors Authors | Massiah, M.A. / Matts, J.A.B. / Short, K.M. / Simmons, B.N. / Singireddy, S. / Zou, J. / Cox, T.C. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2007 Journal: J.Mol.Biol. / Year: 2007Title: Solution Structure of the MID1 B-box2 CHC(D/C)C(2)H(2) Zinc-binding Domain: Insights into an Evolutionarily Conserved RING Fold Authors: Massiah, M.A. / Matts, J.A.B. / Short, K.M. / Simmons, B.N. / Singireddy, S. / Yi, Z. / Cox, T.C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2dq5.cif.gz 2dq5.cif.gz | 270 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2dq5.ent.gz pdb2dq5.ent.gz | 227.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2dq5.json.gz 2dq5.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2dq5_validation.pdf.gz 2dq5_validation.pdf.gz | 348.3 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2dq5_full_validation.pdf.gz 2dq5_full_validation.pdf.gz | 444.5 KB | Display | |
| Data in XML |  2dq5_validation.xml.gz 2dq5_validation.xml.gz | 12.6 KB | Display | |
| Data in CIF |  2dq5_validation.cif.gz 2dq5_validation.cif.gz | 20.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dq/2dq5 https://data.pdbj.org/pub/pdb/validation_reports/dq/2dq5 ftp://data.pdbj.org/pub/pdb/validation_reports/dq/2dq5 ftp://data.pdbj.org/pub/pdb/validation_reports/dq/2dq5 | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 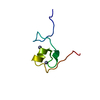
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 5390.228 Da / Num. of mol.: 1 / Fragment: Bbox2 domain Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: MID1 / Plasmid: pGEX-4T2 / Species (production host): Escherichia coli / Production host: Homo sapiens (human) / Gene: MID1 / Plasmid: pGEX-4T2 / Species (production host): Escherichia coli / Production host:  References: UniProt: O15344, Ligases; Forming carbon-nitrogen bonds; Acid-amino-acid ligases (peptide synthases) |
|---|---|
| #2: Chemical |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 0.75-1.0mM 15N & 15N/13C labeled BBOX2; 50mM Tris-HCL, 100mM NaCl, 1mM ZnCl2, 10mM beta-mercaptoethanol; 2% sodium azide; 90% H2O, 10% D2O Solvent system: 90% H2O/10% D2O |
|---|---|
| Sample conditions | Ionic strength: 50mM Tris-HCl; 100mM NaCl; 5mM ZnCl2, 10mM beta-mercaptoethanol pH: 7.5 / Pressure: ambient / Temperature: 294 K |
-NMR measurement
| NMR spectrometer | Type: Varian INOVA / Manufacturer: Varian / Model: INOVA / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: torsion angle dynamics, low target function / Software ordinal: 1 Details: A total of 200 random structures were calculated with CYANA2.1 using a fast torsion angle dynamics algorithm and the best structures were selected based on their low target function (less 1) ...Details: A total of 200 random structures were calculated with CYANA2.1 using a fast torsion angle dynamics algorithm and the best structures were selected based on their low target function (less 1), no NOE violation less than 0.2 Ang, vdw < 0.25, dihedral <3o. | ||||||||||||||||||||
| NMR representative | Selection criteria: minimized average structure, lowest energy, fewest violations, closest to the average | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 200 / Conformers submitted total number: 16 |
 Movie
Movie Controller
Controller








 PDBj
PDBj






 HSQC
HSQC